Deep-Learning-Driven Quantification of Interstitial Fibrosis in Digitized Kidney Biopsies
- PMID: 34033750
- PMCID: PMC8453248
- DOI: 10.1016/j.ajpath.2021.05.005
Deep-Learning-Driven Quantification of Interstitial Fibrosis in Digitized Kidney Biopsies
Abstract
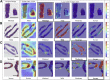
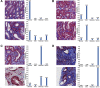
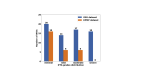
Interstitial fibrosis and tubular atrophy (IFTA) on a renal biopsy are strong indicators of disease chronicity and prognosis. Techniques that are typically used for IFTA grading remain manual, leading to variability among pathologists. Accurate IFTA estimation using computational techniques can reduce this variability and provide quantitative assessment. Using trichrome-stained whole-slide images (WSIs) processed from human renal biopsies, we developed a deep-learning framework that captured finer pathologic structures at high resolution and overall context at the WSI level to predict IFTA grade. WSIs (n = 67) were obtained from The Ohio State University Wexner Medical Center. Five nephropathologists independently reviewed them and provided fibrosis scores that were converted to IFTA grades: ≤10% (none or minimal), 11% to 25% (mild), 26% to 50% (moderate), and >50% (severe). The model was developed by associating the WSIs with the IFTA grade determined by majority voting (reference estimate). Model performance was evaluated on WSIs (n = 28) obtained from the Kidney Precision Medicine Project. There was good agreement on the IFTA grading between the pathologists and the reference estimate (κ = 0.622 ± 0.071). The accuracy of the deep-learning model was 71.8% ± 5.3% on The Ohio State University Wexner Medical Center and 65.0% ± 4.2% on Kidney Precision Medicine Project data sets. Our approach to analyzing microscopic- and WSI-level changes in renal biopsies attempts to mimic the pathologist and provides a regional and contextual estimation of IFTA. Such methods can assist clinicopathologic diagnosis.
Copyright © 2021 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures








References
-
- Amann K., Haas C.S. What you should know about the work-up of a renal biopsy. Nephrol Dial Transpl. 2006;21:1157–1161. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

