Super.FELT: supervised feature extraction learning using triplet loss for drug response prediction with multi-omics data
- PMID: 34034645
- PMCID: PMC8152321
- DOI: 10.1186/s12859-021-04146-z
Super.FELT: supervised feature extraction learning using triplet loss for drug response prediction with multi-omics data
Abstract
Background: Predicting the drug response of a patient is important for precision oncology. In recent studies, multi-omics data have been used to improve the prediction accuracy of drug response. Although multi-omics data are good resources for drug response prediction, the large dimension of data tends to hinder performance improvement. In this study, we aimed to develop a new method, which can effectively reduce the large dimension of data, based on the supervised deep learning model for predicting drug response.
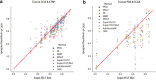
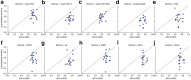
Results: We proposed a novel method called Supervised Feature Extraction Learning using Triplet loss (Super.FELT) for drug response prediction. Super.FELT consists of three stages, namely, feature selection, feature encoding using a supervised method, and binary classification of drug response (sensitive or resistant). We used multi-omics data including mutation, copy number aberration, and gene expression, and these were obtained from cell lines [Genomics of Drug Sensitivity in Cancer (GDSC), Cancer Cell Line Encyclopedia (CCLE), and Cancer Therapeutics Response Portal (CTRP)], patient-derived tumor xenografts (PDX), and The Cancer Genome Atlas (TCGA). GDSC was used for training and cross-validation tests, and CCLE, CTRP, PDX, and TCGA were used for external validation. We performed ablation studies for the three stages and verified that the use of multi-omics data guarantees better performance of drug response prediction. Our results verified that Super.FELT outperformed the other methods at external validation on PDX and TCGA and was good at cross-validation on GDSC and external validation on CCLE and CTRP. In addition, through our experiments, we confirmed that using multi-omics data is useful for external non-cell line data.
Conclusion: By separating the three stages, Super.FELT achieved better performance than the other methods. Through our results, we found that it is important to train encoders and a classifier independently, especially for external test on PDX and TCGA. Moreover, although gene expression is the most powerful data on cell line data, multi-omics promises better performance for external validation on non-cell line data than gene expression data. Source codes of Super.FELT are available at https://github.com/DMCB-GIST/Super.FELT .
Keywords: Drug response prediction; Multi-omics data; Pharmacogenomics; Precision oncology; Triplet loss; encoder using supervised methods.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(1362–4962):955–61. doi: 10.1093/nar/gks1111. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

