Fusion protein EWS-FLI1 is incorporated into a protein granule in cells
- PMID: 34035145
- PMCID: PMC8284321
- DOI: 10.1261/rna.078827.121
Fusion protein EWS-FLI1 is incorporated into a protein granule in cells
Abstract
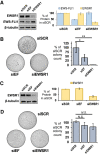
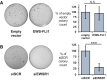
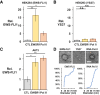
Ewing sarcoma is driven by fusion proteins containing a low complexity (LC) domain that is intrinsically disordered and a powerful transcriptional regulator. The most common fusion protein found in Ewing sarcoma, EWS-FLI1, takes its LC domain from the RNA-binding protein EWSR1 (Ewing Sarcoma RNA-binding protein 1) and a DNA-binding domain from the transcription factor FLI1 (Friend Leukemia Virus Integration 1). EWS-FLI1 can bind RNA polymerase II (RNA Pol II) and self-assemble through its low-complexity (LC) domain. The ability of RNA-binding proteins like EWSR1 to self-assemble or phase separate in cells has raised questions about the contribution of this process to EWS-FLI1 activity. We examined EWSR1 and EWS-FLI1 activity in Ewing sarcoma cells by siRNA-mediated knockdown and RNA-seq analysis. More transcripts were affected by the EWSR1 knockdown than expected and these included many EWS-FLI1 regulated genes. We reevaluated physical interactions between EWS-FLI1, EWSR1, and RNA Pol II, and employed a cross-linking based strategy to investigate protein assemblies associated with the proteins. The LC domain of EWS-FLI1 was required for the assemblies observed to form in cells. These results offer new insights into a protein assembly that may enable EWS-FLI1 to bind its wide network of protein partners and contribute to regulation of gene expression in Ewing sarcoma.
Keywords: Ewing sarcoma; fusion proteins; granules; phase separation; transcription.
Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
Pharmaceutical Interference of the EWS-FLI1-driven Transcriptome By Cotargeting H3K27ac and RNA Polymerase Activity in Ewing Sarcoma.Mol Cancer Ther. 2021 Oct;20(10):1868-1879. doi: 10.1158/1535-7163.MCT-20-0489. Epub 2021 Jul 26. Mol Cancer Ther. 2021. PMID: 34315769
-
Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein.Nat Genet. 1999 Oct;23(2):222-7. doi: 10.1038/13854. Nat Genet. 1999. PMID: 10508522
-
BET bromodomain inhibitors suppress EWS-FLI1-dependent transcription and the IGF1 autocrine mechanism in Ewing sarcoma.Oncotarget. 2016 Jul 12;7(28):43504-43517. doi: 10.18632/oncotarget.9762. Oncotarget. 2016. PMID: 27259270 Free PMC article.
-
Epigenetic and Transcriptional Signaling in Ewing Sarcoma-Disease Etiology and Therapeutic Opportunities.Biomedicines. 2022 Jun 5;10(6):1325. doi: 10.3390/biomedicines10061325. Biomedicines. 2022. PMID: 35740349 Free PMC article. Review.
-
Regulation of Metastasis in Ewing Sarcoma.Cancers (Basel). 2022 Oct 7;14(19):4902. doi: 10.3390/cancers14194902. Cancers (Basel). 2022. PMID: 36230825 Free PMC article. Review.
Cited by
-
Evaluating the local expression pattern of IGF-1R in tumor tissues and the circulating levels of IGF-1, IGFBP-1, and IGFBP-3 in the blood of patients with different primary bone tumors.Front Oncol. 2023 Jan 13;12:1096438. doi: 10.3389/fonc.2022.1096438. eCollection 2022. Front Oncol. 2023. PMID: 36713521 Free PMC article.
-
A Proximity biotinylation assay with a host protein bait reveals multiple factors modulating enterovirus replication.PLoS Pathog. 2022 Oct 28;18(10):e1010906. doi: 10.1371/journal.ppat.1010906. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36306280 Free PMC article.
-
Liquid-liquid phase separation in diseases.MedComm (2020). 2024 Jul 13;5(7):e640. doi: 10.1002/mco2.640. eCollection 2024 Jul. MedComm (2020). 2024. PMID: 39006762 Free PMC article. Review.
-
Molecular features driving condensate formation and gene expression by the BRD4-NUT fusion oncoprotein are overlapping but distinct.Sci Rep. 2023 Jul 24;13(1):11907. doi: 10.1038/s41598-023-39102-9. Sci Rep. 2023. PMID: 37488172 Free PMC article.
-
Regulation of EWSR1-FLI1 Function by Post-Transcriptional and Post-Translational Modifications.Cancers (Basel). 2023 Jan 6;15(2):382. doi: 10.3390/cancers15020382. Cancers (Basel). 2023. PMID: 36672331 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
