Estimating infectiousness throughout SARS-CoV-2 infection course
- PMID: 34035154
- PMCID: PMC9267347
- DOI: 10.1126/science.abi5273
Estimating infectiousness throughout SARS-CoV-2 infection course
Abstract
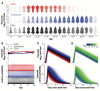
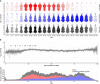
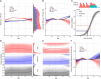
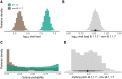
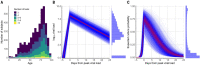
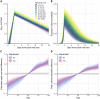
Two elementary parameters for quantifying viral infection and shedding are viral load and whether samples yield a replicating virus isolate in cell culture. We examined 25,381 cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Germany, including 6110 from test centers attended by presymptomatic, asymptomatic, and mildly symptomatic (PAMS) subjects, 9519 who were hospitalized, and 1533 B.1.1.7 lineage infections. The viral load of the youngest subjects was lower than that of the older subjects by 0.5 (or fewer) log10 units, and they displayed an estimated ~78% of the peak cell culture replication probability; in part this was due to smaller swab sizes and unlikely to be clinically relevant. Viral loads above 109 copies per swab were found in 8% of subjects, one-third of whom were PAMS, with a mean age of 37.6 years. We estimate 4.3 days from onset of shedding to peak viral load (108.1 RNA copies per swab) and peak cell culture isolation probability (0.75). B.1.1.7 subjects had mean log10 viral load 1.05 higher than that of non-B.1.1.7 subjects, and the estimated cell culture replication probability of B.1.1.7 subjects was higher by a factor of 2.6.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
References
-
- Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., Park S. Y., Son H.-J., Yu S., Park J. W., Choo E. J., Park S., Loeb M., Kim T. H., Clinical Course and Molecular Viral Shedding Among Asymptomatic and Symptomatic Patients With SARS-CoV-2 Infection in a Community Treatment Center in the Republic of Korea. JAMA Intern. Med. 180, 1447–1452 (2020). 10.1001/jamainternmed.2020.3862 - DOI - PMC - PubMed
-
- Szablewski C. M., Chang K. T., Brown M. M., Chu V. T., Yousaf A. R., Anyalechi N., Aryee P. A., Kirking H. L., Lumsden M., Mayweather E., McDaniel C. J., Montierth R., Mohammed A., Schwartz N. G., Shah J. A., Tate J. E., Dirlikov E., Drenzek C., Lanzieri T. M., Stewart R. J., SARS-CoV-2 Transmission and Infection Among Attendees of an Overnight Camp - Georgia, June 2020. MMWR Morb. Mortal. Wkly. Rep. 69, 1023–1025 (2020). 10.15585/mmwr.mm6931e1 - DOI - PMC - PubMed
-
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L., Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 26, 1200–1204 (2020). 10.1038/s41591-020-0965-6 - DOI - PubMed
-
- Bi Q., Wu Y., Mei S., Ye C., Zou X., Zhang Z., Liu X., Wei L., Truelove S. A., Zhang T., Gao W., Cheng C., Tang X., Wu X., Wu Y., Sun B., Huang S., Sun Y., Zhang J., Ma T., Lessler J., Feng T., Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: A retrospective cohort study. Lancet Infect. Dis. 20, 911–919 (2020). 10.1016/S1473-3099(20)30287-5 - DOI - PMC - PubMed
-
- Waterfield T., Watson C., Moore R., Ferris K., Tonry C., Watt A., McGinn C., Foster S., Evans J., Lyttle M. D., Ahmad S., Ladhani S., Corr M., McFetridge L., Mitchell H., Brown K., Amirthalingam G., Maney J.-A., Christie S., Seroprevalence of SARS-CoV-2 antibodies in children: A prospective multicentre cohort study. Arch. Dis. Child. 10.1136/archdischild-2020-320558 (2020). 10.1136/archdischild-2020-320558 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

