Complement C1s cleaves PfEMP1 at interdomain conserved sites inhibiting Plasmodium falciparum cytoadherence
- PMID: 34035177
- PMCID: PMC8179237
- DOI: 10.1073/pnas.2104166118
Complement C1s cleaves PfEMP1 at interdomain conserved sites inhibiting Plasmodium falciparum cytoadherence
Abstract
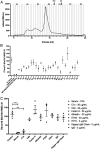
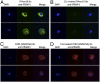
Cytoadhesion of Plasmodium falciparum-infected erythrocytes (IEs) to the endothelial lining of blood vessels protects parasites from splenic destruction, but also leads to detrimental inflammation and vessel occlusion. Surface display of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) adhesion ligands exposes them to host antibodies and serum proteins. PfEMP1 are important targets of acquired immunity to malaria, and through evolution, the protein family has expanded and diversified to bind a select set of host receptors through antigenically diversified receptor-binding domains. Here, we show that complement component 1s (C1s) in serum cleaves PfEMP1 at semiconserved arginine motifs located at interdomain regions between the receptor-binding domains, rendering the IE incapable of binding the two main PfEMP1 receptors, CD36 and endothelial protein C receptor (EPCR). Bioinformatic analyses of PfEMP1 protein sequences from 15 P. falciparum genomes found the C1s motif was present in most PfEMP1 variants. Prediction of C1s cleavage and loss of binding to endothelial receptors was further corroborated by testing of several different parasite lines. These observations suggest that the parasites have maintained susceptibility for cleavage by the serine protease, C1s, and provides evidence for a complex relationship between the complement system and the P. falciparum cytoadhesion virulence determinant.
Keywords: C1s; EPCR; PfEMP1; cytoadhesion; malaria.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- World Health Organization , “World malaria report 2020: 20 years of global progress and challenges” (World Health Organization, Geneva, 2020).
-
- Idro R., Jenkins N. E., Newton C. R., Pathogenesis, clinical features, and neurological outcome of cerebral malaria. Lancet Neurol. 4, 827–840 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

