Decreased GLUT2 and glucose uptake contribute to insulin secretion defects in MODY3/HNF1A hiPSC-derived mutant β cells
- PMID: 34035238
- PMCID: PMC8149827
- DOI: 10.1038/s41467-021-22843-4
Decreased GLUT2 and glucose uptake contribute to insulin secretion defects in MODY3/HNF1A hiPSC-derived mutant β cells
Abstract
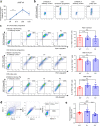
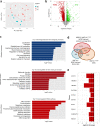
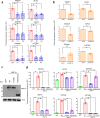
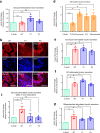
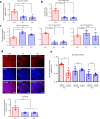
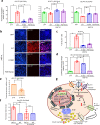
Heterozygous HNF1A gene mutations can cause maturity onset diabetes of the young 3 (MODY3), characterized by insulin secretion defects. However, specific mechanisms of MODY3 in humans remain unclear due to lack of access to diseased human pancreatic cells. Here, we utilize MODY3 patient-derived human induced pluripotent stem cells (hiPSCs) to study the effect(s) of a causal HNF1A+/H126D mutation on pancreatic function. Molecular dynamics simulations predict that the H126D mutation could compromise DNA binding and gene target transcription. Genome-wide RNA-Seq and ChIP-Seq analyses on MODY3 hiPSC-derived endocrine progenitors reveal numerous HNF1A gene targets affected by the mutation. We find decreased glucose transporter GLUT2 expression, which is associated with reduced glucose uptake and ATP production in the MODY3 hiPSC-derived β-like cells. Overall, our findings reveal the importance of HNF1A in regulating GLUT2 and several genes involved in insulin secretion that can account for the insulin secretory defect clinically observed in MODY3 patients.
Conflict of interest statement
N.H.J.N. and A.K.K.T. are co-founders of BetaLife Pte Ltd. C.S.V. is the co-founder of Sinopsee Therapeutics and Aplomex. The other authors declare no competing interests.
Figures


References
-
- Mendel DB, Crabtree GR. HNF-1, a member of a novel proteins. Biochemistry. 1991;266:677–680. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

