Control of mouse limb initiation and antero-posterior patterning by Meis transcription factors
- PMID: 34035267
- PMCID: PMC8149412
- DOI: 10.1038/s41467-021-23373-9
Control of mouse limb initiation and antero-posterior patterning by Meis transcription factors
Abstract
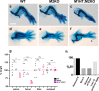
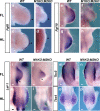
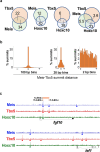
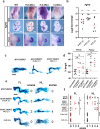
Meis1 and Meis2 are homeodomain transcription factors that regulate organogenesis through cooperation with Hox proteins. Elimination of Meis genes after limb induction has shown their role in limb proximo-distal patterning; however, limb development in the complete absence of Meis function has not been studied. Here, we report that Meis1/2 inactivation in the lateral plate mesoderm of mouse embryos leads to limb agenesis. Meis and Tbx factors converge in this function, extensively co-binding with Tbx to genomic sites and co-regulating enhancers of Fgf10, a critical factor in limb initiation. Limbs with three deleted Meis alleles show proximal-specific skeletal hypoplasia and agenesis of posterior skeletal elements. This failure in posterior specification results from an early role of Meis factors in establishing the limb antero-posterior prepattern required for Shh activation. Our results demonstrate roles for Meis transcription factors in early limb development and identify their involvement in previously undescribed interaction networks that regulate organogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

