Gut microbiota composition in health-care facility-and community-onset diarrheic patients with Clostridioides difficile infection
- PMID: 34035404
- PMCID: PMC8149855
- DOI: 10.1038/s41598-021-90380-7
Gut microbiota composition in health-care facility-and community-onset diarrheic patients with Clostridioides difficile infection
Abstract
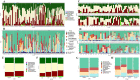
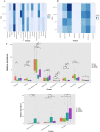
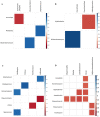
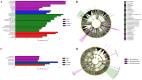
The role of gut microbiota in the establishment and development of Clostridioides difficile infection (CDI) has been widely discussed. Studies showed the impact of CDI on bacterial communities and the importance of some genera and species in recovering from and preventing infection. However, most studies have overlooked important components of the intestinal ecosystem, such as eukaryotes and archaea. We investigated the bacterial, archaea, and eukaryotic intestinal microbiota of patients with health-care-facility- or community-onset (HCFO and CO, respectively) diarrhea who were positive or negative for CDI. The CDI-positive groups (CO/+, HCFO/+) showed an increase in microorganisms belonging to Bacteroidetes, Firmicutes, Proteobacteria, Ascomycota, and Opalinata compared with the CDI-negative groups (CO/-, HCFO/-). Patients with intrahospital-acquired diarrhea (HCFO/+, HCFO/-) showed a marked decrease in bacteria beneficial to the intestine, and there was evidence of increased Archaea and Candida and Malassezia species compared with the CO groups (CO/+, CO/-). Characteristic microbiota biomarkers were established for each group. Finally, correlations between bacteria and eukaryotes indicated interactions among the different kingdoms making up the intestinal ecosystem. We showed the impact of CDI on microbiota and how it varies with where the infection is acquired, being intrahospital-acquired diarrhea one of the most influential factors in the modulation of bacterial, archaea, and eukaryotic populations. We also highlight interactions between the different kingdoms of the intestinal ecosystem, which need to be evaluated to improve our understanding of CDI pathophysiology.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

