Base-edited CAR T cells for combinational therapy against T cell malignancies
- PMID: 34035409
- PMCID: PMC8632682
- DOI: 10.1038/s41375-021-01282-6
Base-edited CAR T cells for combinational therapy against T cell malignancies
Abstract
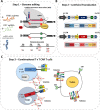
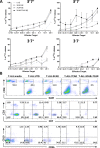
Targeting T cell malignancies using chimeric antigen receptor (CAR) T cells is hindered by 'T v T' fratricide against shared antigens such as CD3 and CD7. Base editing offers the possibility of seamless disruption of gene expression of problematic antigens through creation of stop codons or elimination of splice sites. We describe the generation of fratricide-resistant T cells by orderly removal of TCR/CD3 and CD7 ahead of lentiviral-mediated expression of CARs specific for CD3 or CD7. Molecular interrogation of base-edited cells confirmed elimination of chromosomal translocations detected in conventional Cas9 treated cells. Interestingly, 3CAR/7CAR co-culture resulted in 'self-enrichment' yielding populations 99.6% TCR-/CD3-/CD7-. 3CAR or 7CAR cells were able to exert specific cytotoxicity against leukaemia lines with defined CD3 and/or CD7 expression as well as primary T-ALL cells. Co-cultured 3CAR/7CAR cells exhibited highest cytotoxicity against CD3 + CD7 + T-ALL targets in vitro and an in vivo human:murine chimeric model. While APOBEC editors can reportedly exhibit guide-independent deamination of both DNA and RNA, we found no problematic 'off-target' activity or promiscuous base conversion affecting CAR antigen-specific binding regions, which may otherwise redirect T cell specificity. Combinational infusion of fratricide-resistant anti-T CAR T cells may enable enhanced molecular remission ahead of allo-HSCT for T cell malignancies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Litzow MR, Ferrando AA. How I treat T-cell acute lymphoblastic leukemia in adults. Blood. 2015;126:833–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

