Long non-coding RNA DUXAP8 promotes tumorigenesis by regulating IGF1R via miR-9-3p in hepatocellular carcinoma
- PMID: 34035852
- PMCID: PMC8135127
- DOI: 10.3892/etm.2021.10187
Long non-coding RNA DUXAP8 promotes tumorigenesis by regulating IGF1R via miR-9-3p in hepatocellular carcinoma
Abstract
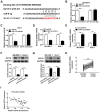
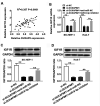
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide with a low 5-year survival rate. Long non-coding RNA (lncRNA) double homeobox A pseudogene 8 (DUXAP8) is an oncogene and a potential biomarker in various tumors, such as ovarian, colorectal and non-small-cell lung cancer. However, the function and molecular mechanism underlying DUXAP8 in HCC progression is not completely understood. The expression of DUXAP8, microRNA (miR)-9-3p and insulin-like growth factor 1 receptor (IGF1R) in HCC tissues and cells was detected via reverse transcription-quantitative PCR. The expression levels of IGF1R and epithelial-mesenchymal transition-associated proteins (Snail, Slug, E-cadherin, N-cadherin and vimentin) were assessed via western blotting. The effects of DUXAP8, miR-9-3p and IGF1R on proliferation, migration and invasion were examined by conducting Cell Counting Kit-8 and Transwell assays, respectively. The interaction between miR-9-3p and DUXAP8 or IGF1R was predicted using StarBase or TargetScan, and further assessed using dual luciferase reporter and RNA immunoprecipitation assays. DUXAP8 and IGF1R were upregulated and miR-9-3p was downregulated in HCC tissues and cells compared with adjacent healthy tissues and a normal liver cell line, respectively. miR-9-3p overexpression decreased the protein expression level of IGF1R, and miR-9-3p knockdown enhanced the protein expression level of IGF1R in HCC cells compared with the corresponding control groups. Moreover, compared with the corresponding control groups, DUXAP8 knockdown and miR-9-3p overexpression increased E-cadherin protein expression levels, and decreased Snail, Slug, N-cadherin and vimentin protein expression levels. However, miR-9-3p inhibitor and IGF1R overexpression reversed DUXAP8 knockdown- and miR-9-3p overexpression-induced effects, respectively. In addition, compared with the corresponding control groups, DUXAP8 knockdown and miR-9-3p overexpression suppressed proliferation, migration and invasion, which was reversed by miR-9-3p inhibitor and IGF1R overexpression, respectively. Moreover, miR-9-3p as the target of DUXAP8 and IGF1R as the target of miR-9-3p were verified in HCC cells. lncRNA DUXAP8 contributed to HCC tumorigenesis via the miR-9-3p/IGF1R axis, providing a novel therapeutic approach for HCC diagnosis and treatment.
Keywords: epithelial-mesenchymal transition; hepatocellular carcinoma; insulin-like growth factor 1 receptor; invasion; long non-coding RNA double homeobox A pseudogene 8; microRNA-9-3p; migration; proliferation.
Copyright: © Guan et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
