The Potential Genes Mediate the Pathogenicity of Allogeneic CD4+T Cell in aGVHD Mouse Model
- PMID: 34036106
- PMCID: PMC8121574
- DOI: 10.1155/2021/9958745
The Potential Genes Mediate the Pathogenicity of Allogeneic CD4+T Cell in aGVHD Mouse Model
Abstract
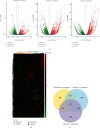
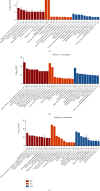
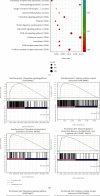
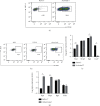
Acute graft-versus-host disease (aGVHD) remains a significant and severe complication of allogeneic hematopoietic stem cell transplantation (allo-HSCT). Due to the occurrence of aGVHD, allo-HSCT significantly increases the mortality rate compared with autologous hematopoietic stem cell transplantation (auto-HSCT). In this study, auto-HSCT and allo-HSCT aGVHD mouse models were built to detect the difference in CD4+ lymphocyte in different tissues based on ribonucleic acid sequencing (RNA-Seq) analysis. Clustering analysis, functional annotation, and pathway enrichment analysis were performed on differentially expressed genes (DEGs). The protein-protein interaction (PPI) network was used to find hub genes. CD4+T cells were activated by MLR and cytokine stimulation. Cells were sorted out by a flow cell sorter. The selected genes were verified by qRT-PCR, histology, and immunofluorescence staining. The GSE126518 GEO dataset was used to verify the hub genes. Enrichment analysis revealed four immune-related pathways that play an important role in aGVHD, including immunoregulatory interactions between a lymphoid and a nonlymphoid cell, chemokine receptors binding chemokines, cytokine and cytokine receptor interaction, and the chemokine signaling pathway. At the same time, with the PPI network, 11 novel hub genes that were most likely to participate in immunoregulation in aGVHD were identified, which were further validated by qRT-PCR and the GSE126518 dataset. Besides, the protein expression level of Cxcl7 was consistent with the sequencing results. In summary, this study revealed that immunoregulation-related DEGs and pathways played a vital role in the onset of aGVHD. These findings may provide some new clues for probing the pathogenesis and treatment of aGVHD.
Copyright © 2021 Zhengyu Yu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

