Impact of a 40-Gene Targeted Panel Test on Physician Decision Making for Patients With Acute Myeloid Leukemia
- PMID: 34036230
- PMCID: PMC8140802
- DOI: 10.1200/PO.20.00182
Impact of a 40-Gene Targeted Panel Test on Physician Decision Making for Patients With Acute Myeloid Leukemia
Abstract
Purpose: Physicians treating hematologic malignancies increasingly order targeted sequencing panels to interrogate recurrently mutated genes. The precise impact of these panels on clinical decision making is not well understood.
Methods: Here, we report our institutional experience with a targeted 40-gene panel (MyeloSeq) that is used to generate a report for both genetic variants and variant allele frequencies for the treating physician (the limit of mutation detection is approximately one AML cell in 50).
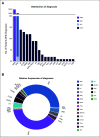
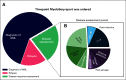
Results: In total, 346 sequencing reports were generated for 325 patients with suspected hematologic malignancies over an 8-month period (August 2018 to April 2019). To determine the influence of genomic data on clinical care for patients with acute myeloid leukemia (AML), we analyzed 122 consecutive reports from 109 patients diagnosed with AML and surveyed the treating physicians with a standardized questionnaire. The panel was ordered most commonly at diagnosis (61.5%), but was also used to assess response to therapy (22.9%) and to detect suspected relapse (15.6%). The panel was ordered at multiple timepoints during the disease course for 11% of patients. Physicians self-reported that 50 of 114 sequencing reports (44%) influenced clinical care decisions in 44 individual patients. Influences were often nuanced and extended beyond identifying actionable genetic variants with US Food and Drug Administration-approved drugs.
Conclusion: This study provides insights into how physicians are currently using multigene panels capable of detecting relatively rare AML cells. The most influential way to integrate these tools into clinical practice will be to perform prospective clinical trials that assess patient outcomes in response to genomically driven interventions.
© 2021 by American Society of Clinical Oncology.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center. Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments). Erica K. BarnellEmployment: Geneoscopy Stock and Other Ownership Interests: Geneoscopy Patents, Royalties, Other Intellectual Property: Inventor of intellectual property in start-up company (Geneoscopy) Travel, Accommodations, Expenses: GeneoscopyKenneth F. NewcomerEmployment: Geneoscopy (I) Stock and Other Ownership Interests: Geneoscopy (I) Patents, Royalties, Other Intellectual Property: Inventor of intellectual property owned by Geneoscopy (I)Zachary L. SkidmoreStock and Other Ownership Interests: Aim Immunotech, Catalyst PharmaceuticalsKilannin KrysiakConsulting or Advisory Role: Gerson Lehrman GroupLukas D. WartmanConsulting or Advisory Role: Novartis, IncyteStephen T. OhConsulting or Advisory Role: Incyte, Novartis, Blueprint Medicines, Celgene, Kartos, CTI BioPharma Corp, PharmaEssentia, Disc Medicine, Constellation Pharmaceuticals Research Funding: Incyte (Inst), Gilead Sciences (Inst), CTI BioPharma Corp (Inst), Kartos (Inst), Celgene (Inst), Sierra Oncology (Inst), Blueprint Medicines (Inst), Constellation Pharmaceuticals (Inst)John S. WelchConsulting or Advisory Role: Archer Biosciences, Agios Research Funding: Janssen Oncology (Inst), Notable Labs (Inst)Keith E. Stockerl-GoldsteinStock and Other Ownership Interests: Abbott Laboratories, AbbVie Consulting or Advisory Role: Celgene Research Funding: GlaxoSmithKline, Takeda, BiolineRx, Janssen Other Relationship: CellerantRavi VijConsulting or Advisory Role: Bristol Myers Squibb, Celgene, Janssen, Sanofi, Karyopharm Therapeutics, Takeda, Genentech, AbbVie, Oncopeptides Research Funding: Takeda, Celgene, Bristol Myers Squibb Travel, Accommodations, Expenses: Celgene, Bristol Myers Squibb, Sanofi, Janssen, DAVAOncology, Karyopharm Therapeutics, Amgen, Takeda, AbbVieAmanda F. CashenConsulting or Advisory Role: Agios Speakers' Bureau: Novartis, CelgeneIskra PusicConsulting or Advisory Role: Kadmon, Incyte, SyndaxCamille N. AbboudStock and Other Ownership Interests: AbbVie (I), Abbott Laboratories (I), Gilead Sciences (I), Bristol Myers Squibb (I), Johnson & Johnson (I) Honoraria: Cardinal Health, Dova Pharmaceuticals, NkartaTherapeutics, Archer Biosciences Research Funding: Actinium Pharmaceuticals, Selvita (Inst), AlloVir (Inst), Forty-Seven (Inst)Armin GhobadiHonoraria: Kite Pharma Consulting or Advisory Role: Kite Pharma, Celgene, Amgen, Wugen Speakers' Bureau: Kite Pharma, AmgenGeoffrey L. UyConsulting or Advisory Role: Astellas Pharma, Genentech, Jazz PharmaceuticalsMark A. SchroederConsulting or Advisory Role: Astellas Pharma, Dova Pharmaceuticals, Flatiron Health, GlaxoSmithKline, Incyte, Partners Therapeutics, Partners Therapeutics, Pfizer Speakers' Bureau: AbbVie, Merck, TakedaJohn F. DiPersioStock and Other Ownership Interests: Magenta Therapeutics, Wugen Honoraria: Incyte Consulting or Advisory Role: Cellworks, Rivervest, Magenta Therapeutics, Incyte Research Funding: Amphivena Therapeutics (Inst), Macrogenics (Inst), Incyte (Inst), Wugen (Inst), BiolineRx (Inst), Maxcyte (Inst), Bigelow Aerospace (Inst) Patents, Royalties, Other Intellectual Property: Patents: CD7 and CD2 knockout for CART to CD7 and CDL; Duvelisib for treatment of cytokine release syndrome; NT-17 to enhance CART survival; Novel WU mobilizing compounds (Inst); Selection of IMPDH mutant stem cells; IFNγ, upregulate MHCII for relapsed AML; Dextran-based molecules to detect CAR-T cells; Combining integrin inhibitor with chemokine binders, 016131; JAK and calcineurin inhibition, solid organ transplant; VLA4, gro-b; Triple combination–CXCR2, VLA-4, gro-b; Targeting IFNR/CSCR3 in GvHD; WU/SLU compounds VLA4 and CXCR2 (Inst) Travel, Accommodations, Expenses: Incyte, Macrogenics, Magenta TherapeuticsMary C. PolitiResearch Funding: MerckDavid H. SpencerConsulting or Advisory Role: Wugen Research Funding: IlluminaEric J. DuncavageStock and Other Ownership Interests: P&V Licensing Consulting or Advisory Role: Cofactor Genomics, Bristol Myers Squibb, Eli Lilly Open Payments Link: https://openpaymentsdata.cms.gov/physician/317379Timothy J. LeyResearch Funding: Rigel Patents, Royalties, Other Intellectual Property: Licenses for monoclonal antibodies and recombinant granzymesMeagan A. JacobyHonoraria: Takeda Research Funding: Geron (Inst), AbbVie (Inst), Jazz Pharmaceuticals (Inst), Aprea AB (Inst), Amphivena (Inst), Janssen (Inst) No other potential conflicts of interest were reported.
Figures



References
-
- Ray T. Survey of precision oncology programs finds agreement on testing, divergence in care delivery. Precision Oncology News; https://www.precisiononcologynews.com/cancer/survey-precision-oncology-p... July 19, 2019.
-
- Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists J Mol Diagn 194–232017 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

