Comprehensive genome-wide analysis of routine non-invasive test data allows cancer prediction: A single-center retrospective analysis of over 85,000 pregnancies
- PMID: 34036251
- PMCID: PMC8138727
- DOI: 10.1016/j.eclinm.2021.100856
Comprehensive genome-wide analysis of routine non-invasive test data allows cancer prediction: A single-center retrospective analysis of over 85,000 pregnancies
Abstract
Background: Implausible false positive results in non-invasive prenatal testing (NIPT) have been occasionally associated with the detection of occult maternal malignancies. Hence, there is a need for approaches allowing accurate prediction of whether the NIPT result is pointing to an underlying malignancy, as well as for organized programs ensuring efficient downstream clinical management of these cases.
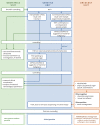
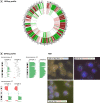
Methods: Using a data set of 88,294 NIPT performed at University Hospital Leuven (Belgium) between November 2013 and March 2020, we retrospectively evaluated the positive predictive value (PPV) of our NIPT approach for cancer detection. In this approach, whole-genome cell-free DNA (cfDNA) data from NIPT were scrutinized for the presence of (sub)chromosomal copy number alterations (CNAs) predictive for a malignancy, using an unbiased NIPT analysis pipeline coined GIPSeq. For suspected cases, the presence of a maternal cancer was evaluated via subsequent multidisciplinary clinical follow-up examinations. The cancer-specificity of the identified CNAs in cfDNA was assessed through genetic analyses of a tumor biopsy.
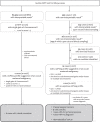
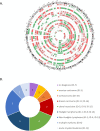
Findings: Fifteen women without a cancer history were identified with a GIPSeq result suggestive of a malignant process. Their cfDNA profiles showed either genome-wide aberrations or a single trisomy 8. Upon clinical examinations, a solid or hematological cancer was identified in 4 and 7 cases, respectively. Three women were identified as having a clonal mosaicism. For one case no underlying condition was found. These numbers add to a PPV of 73%. Based on this experience, we presented a multidisciplinary care path for efficient clinical management of these cases.
Interpretation: The presented approach for analysing NIPT results has a high PPV, yet unknown sensitivity, for detecting asymptomatic malignancies upon routine NIPT. Given the complexity of diagnosing a pregnant woman with cancer, clinical follow-up should occur in a well-designed multidisciplinary setting, such as via the care model that we presented here.
Funding: This work was supported by Research Foundation Flanders and KU Leuven funding.
Keywords: Cancer detection; Clinical follow-up; Non-invasive prenatal testing.
© 2021 The Author(s).
Conflict of interest statement
EL reports personal fees from Springworks Therapeutics outside the submitted work. All other authors declare no competing interests.
Figures
Similar articles
-
Breast Cancer Detection and Treatment Monitoring Using a Noninvasive Prenatal Testing Platform: Utility in Pregnant and Nonpregnant Populations.Clin Chem. 2020 Nov 1;66(11):1414-1423. doi: 10.1093/clinchem/hvaa196. Clin Chem. 2020. PMID: 33141904
-
Clinical utility of non-invasive prenatal testing in pregnancies with ultrasound anomalies.Ultrasound Obstet Gynecol. 2017 Jun;49(6):721-728. doi: 10.1002/uog.17228. Ultrasound Obstet Gynecol. 2017. PMID: 27515011 Free PMC article.
-
Fetal fraction of cell-free DNA in pregnancies after fresh or frozen embryo transfer following assisted reproductive technologies.Hum Reprod. 2020 Jun 1;35(6):1267-1275. doi: 10.1093/humrep/deaa110. Hum Reprod. 2020. PMID: 32539141
-
Non-invasive prenatal testing for fetal chromosomal abnormalities by low-coverage whole-genome sequencing of maternal plasma DNA: review of 1982 consecutive cases in a single center.Ultrasound Obstet Gynecol. 2014 Mar;43(3):254-64. doi: 10.1002/uog.13277. Epub 2014 Feb 10. Ultrasound Obstet Gynecol. 2014. PMID: 24339153 Review.
-
Predicting fetoplacental mosaicism during cfDNA-based NIPT.Curr Opin Obstet Gynecol. 2020 Apr;32(2):152-158. doi: 10.1097/GCO.0000000000000610. Curr Opin Obstet Gynecol. 2020. PMID: 31977337 Review.
Cited by
-
Pushing the boundaries. Concurrent Hodgkin lymphoma and breast cancer treatment with preservation of pregnancy: A case report.Gynecol Oncol Rep. 2022 Jan 31;39:100937. doi: 10.1016/j.gore.2022.100937. eCollection 2022 Feb. Gynecol Oncol Rep. 2022. PMID: 35146105 Free PMC article.
-
Dying To Find Out: The Cost of Time at the Dawn of the Multicancer Early Detection Era.Cancer Epidemiol Biomarkers Prev. 2023 Aug 1;32(8):1003-1010. doi: 10.1158/1055-9965.EPI-22-1275. Cancer Epidemiol Biomarkers Prev. 2023. PMID: 37255363 Free PMC article. Review.
-
Copy-number alterations in cell-free DNA can be transient or harbingers of clonal hematopoiesis.NPJ Precis Oncol. 2025 Mar 25;9(1):88. doi: 10.1038/s41698-025-00877-x. NPJ Precis Oncol. 2025. PMID: 40133611 Free PMC article.
-
Machine learning-based detection of immune-mediated diseases from genome-wide cell-free DNA sequencing datasets.NPJ Genom Med. 2022 Sep 14;7(1):55. doi: 10.1038/s41525-022-00325-w. NPJ Genom Med. 2022. PMID: 36100603 Free PMC article.
-
Initial Clinical Experience with NIPT for Rare Autosomal Aneuploidies and Large Copy Number Variations.J Clin Med. 2022 Jan 13;11(2):372. doi: 10.3390/jcm11020372. J Clin Med. 2022. PMID: 35054066 Free PMC article.
References
-
- Samura O. Update on noninvasive prenatal testing: a review based on current worldwide research. J Obstet Gynaecol Res. 2020;46:1246–1254. - PubMed
-
- Brady P., Brison N., Van Den Bogaert K. Clinical implementation of NIPT - technical and biological challenges. Clin Genet. 2016;89:523–530. - PubMed
-
- Ji X., Li J., Huang Y. Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet Med. 2019;21:2293–2302. - PubMed
-
- Dharajiya N.G., Grosu D.S., Farkas D.H. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin Chem. 2018;64:329–335. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

