Invariant natural killer T-cell subsets have diverse graft-versus-host-disease-preventing and antitumor effects
- PMID: 34036317
- PMCID: PMC8432044
- DOI: 10.1182/blood.2021010887
Invariant natural killer T-cell subsets have diverse graft-versus-host-disease-preventing and antitumor effects
Abstract
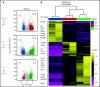
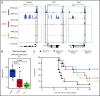
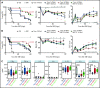
Invariant natural killer T (iNKT) cells are a T-cell subset with potent immunomodulatory properties. Experimental evidence in mice and observational studies in humans indicate that iNKT cells have antitumor potential as well as the ability to suppress acute and chronic graft-versus-host-disease (GVHD). Murine iNKT cells differentiate during thymic development into iNKT1, iNKT2, and iNKT17 sublineages, which differ transcriptomically and epigenomically and have subset-specific developmental requirements. Whether distinct iNKT sublineages also differ in their antitumor effect and their ability to suppress GVHD is currently unknown. In this work, we generated highly purified murine iNKT sublineages, characterized their transcriptomic and epigenomic landscape, and assessed specific functions. We show that iNKT2 and iNKT17, but not iNKT1, cells efficiently suppress T-cell activation in vitro and mitigate murine acute GVHD in vivo. Conversely, we show that iNKT1 cells display the highest antitumor activity against murine B-cell lymphoma cells both in vitro and in vivo. Thus, we report for the first time that iNKT sublineages have distinct and different functions, with iNKT1 cells having the highest antitumor activity and iNKT2 and iNKT17 cells having immune-regulatory properties. These results have important implications for the translation of iNKT cell therapies to the clinic for cancer immunotherapy as well as for the prevention and treatment of GVHD.
© 2021 by The American Society of Hematology.
Figures





Comment in
-
The iNKT-dependent butterfly effect in HCT.Blood. 2021 Sep 9;138(10):821-822. doi: 10.1182/blood.2021012346. Blood. 2021. PMID: 34499153 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

