Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency
- PMID: 34036344
- PMCID: PMC8554652
- DOI: 10.1182/blood.2020007401
Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency
Erratum in
-
Wang Y, Li W, Schulz VP, et al. Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. Blood. 2021;138(17):1615-1627.Blood. 2022 Mar 10;139(10):1600. doi: 10.1182/blood.2021015264. Blood. 2022. PMID: 35267000 Free PMC article. No abstract available.
Abstract
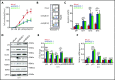
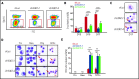
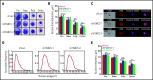
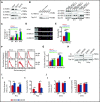
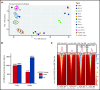
Histone deacetylases (HDACs) are a group of enzymes that catalyze the removal of acetyl groups from histone and nonhistone proteins. HDACs have been shown to have diverse functions in a wide range of biological processes. However, their roles in mammalian erythropoiesis remain to be fully defined. This study showed that, of the 11 classic HDAC family members, 6 (HDAC1, -2, -3, and HDAC5, -6, -7) are expressed in human erythroid cells, with HDAC5 most significantly upregulated during terminal erythroid differentiation. Knockdown of HDAC5 by either short hairpin RNA or small interfering RNA in human CD34+ cells followed by erythroid cell culture led to increased apoptosis, decreased chromatin condensation, and impaired enucleation of erythroblasts. Biochemical analyses revealed that HDAC5 deficiency resulted in activation of p53 in association with increased acetylation of p53. Furthermore, although acetylation of histone 4 (H4) is decreased during normal terminal erythroid differentiation, HDAC5 deficiency led to increased acetylation of H4 (K12) in late-stage erythroblasts. This increased acetylation was accompanied by decreased chromatin condensation, implying a role for H4 (K12) deacetylation in chromatin condensation. ATAC-seq and RNA sequencing analyses revealed that HDAC5 knockdown leads to increased chromatin accessibility genome-wide and global changes in gene expression. Moreover, pharmacological inhibition of HDAC5 by the inhibitor LMK235 also led to increased H4 acetylation, impaired chromatin condensation, and enucleation. Taken together, our findings have uncovered previously unrecognized roles and molecular mechanisms of action for HDAC5 in human erythropoiesis. These results may provide insights into understanding the anemia associated with HDAC inhibitor treatment.
© 2021 by The American Society of Hematology.
Figures


References
-
- D’Andrea AD, Lodish HF, Wong GG. Expression cloning of the murine erythropoietin receptor. Cell. 1989;57(2):277-285. - PubMed
-
- Koury MJ, Bondurant MC. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science. 1990;248(4953):378-381. - PubMed
-
- Lin CS, Lim SK, D’Agati V, Costantini F. Differential effects of an erythropoietin receptor gene disruption on primitive and definitive erythropoiesis. Genes Dev. 1996;10(2):154-164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

