A novel expressed prostatic secretion (EPS)-urine metabolomic signature for the diagnosis of clinically significant prostate cancer
- PMID: 34037347
- PMCID: PMC8185872
- DOI: 10.20892/j.issn.2095-3941.2020.0617
A novel expressed prostatic secretion (EPS)-urine metabolomic signature for the diagnosis of clinically significant prostate cancer
Abstract
Objective: Significant efforts are currently being made to identify novel biomarkers for the diagnosis and risk stratification of prostate cancer (PCa). Metabolomics can be a very useful approach in biomarker discovery because metabolites are an important read-out of the disease when characterized in biological samples. We aimed to determine a metabolomic signature which can accurately distinguish men with clinically significant PCa from those affected by benign prostatic hyperplasia (BPH).
Methods: We first performed untargeted metabolomics using ultrahigh-performance liquid chromatography tandem mass spectrometry on expressed prostatic secretion urine (EPS-urine) from 25 patients affected by BPH and 25 men with clinically significant PCa (defined as Gleason score ≥ 3 + 4). Diagnosis was histologically confirmed after surgical treatment. The EPS-urine metabolomic approach was then applied to a larger, prospective cohort of 92 consecutive patients undergoing multiparametric magnetic resonance imaging for clinical suspicion of PCa prior to biopsy.
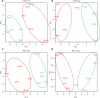
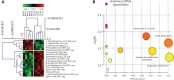
Results: We established a novel metabolomic signature capable of accurately distinguishing PCa from benign tissue. A metabolomic signature was associated with clinically significant PCa in all subgroups of the Prostate Imaging Reporting and Data System (PI-RADS) classification (100% and 89.13% of accuracy when the PI-RADS was in range of 1-2 and 4-5, respectively, and 87.50% in the more critical cases when the PI-RADS was 3).
Conclusions: A combination of metabolites and clinical variables can effectively help in identifying PCa patients that might be overlooked by current imaging technologies. Metabolites from EPS-urine should help in defining the diagnostic pathway of PCa, thus improving PCa detection and decreasing the number of unnecessary prostate biopsies.
Keywords: EPS-urine; Prostate; cancer; diagnosis; metabolomics; prediction.
Copyright © 2021 Cancer Biology & Medicine.
Conflict of interest statement
No potential conflicts of interest are disclosed.
Figures

References
-
- Cooperberg MR, Carroll PR, Klotz L. Active surveillance for prostate cancer: progress and promise. J Clin Oncol. 2011;29:3669–76. - PubMed
-
- Cucchiara V, Cooperberg MR, Dall’Era M, Lin DW, Montorsi F, Schalken JA, et al. Genomic markers in prostate cancer decision making. Eur Urol. 2018;73:572–82. - PubMed
-
- Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–22. - PubMed
-
- Lucarelli G, Loizzo D, Ferro M, Rutigliano M, Vartolomei MD, Cantiello F, et al. Metabolomic profiling for the identification of novel diagnostic markers and therapeutic targets in prostate cancer: an update. Expert Rev Mol Diagn. 2019;19:377–87. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
