An Amino Acid Polymorphism within the HIV-1 Nef Dileucine Motif Functionally Uncouples Cell Surface CD4 and SERINC5 Downregulation
- PMID: 34037423
- PMCID: PMC8312866
- DOI: 10.1128/JVI.00588-21
An Amino Acid Polymorphism within the HIV-1 Nef Dileucine Motif Functionally Uncouples Cell Surface CD4 and SERINC5 Downregulation
Abstract
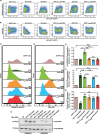
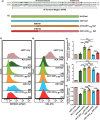
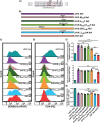
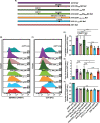
Serine incorporator 5 (SERINC5) reduces the infectivity of progeny HIV-1 virions by incorporating into the outer host-derived viral membrane during egress. To counter SERINC5, the HIV-1 accessory protein Nef triggers SERINC5 internalization by engaging the adaptor protein 2 (AP-2) complex using the [D/E]xxxL[L/I]167 Nef dileucine motif. Nef also engages AP-2 via its dileucine motif to downregulate the CD4 receptor. Although these two Nef functions are related, the mechanisms governing SERINC5 downregulation are incompletely understood. Here, we demonstrate that two primary Nef isolates, referred to as 2410 and 2391 Nef, acquired from acutely HIV-1 infected women from Zimbabwe, both downregulate CD4 from the cell surface. However, only 2410 Nef retains the ability to downregulate cell surface SERINC5. Using a series of Nef chimeras, we mapped the region of 2391 Nef responsible for the functional uncoupling of these two antagonistic pathways to the dileucine motif. Modifications of the first and second x positions of the 2410 Nef dileucine motif to asparagine and aspartic acid residues, respectively (ND164), impaired cell surface SERINC5 downregulation, which resulted in reduced infectious virus yield in the presence of SERINC5. The ND164 mutation additionally partially impaired, but did not completely abrogate, Nef-mediated cell surface CD4 downregulation. Furthermore, the patient infected with HIV-1 encoding 2391 Nef had stable CD4+ T cell counts, whereas infection with HIV-1 encoding 2410 Nef resulted in CD4+ T cell decline and disease progression. IMPORTANCE A contributing factor to HIV-1 persistence is evasion of the host immune response. HIV-1 uses the Nef accessory protein to evade the antiviral roles of the adaptive and intrinsic innate immune responses. Nef targets SERINC5, a restriction factor which potently impairs HIV-1 infection by triggering SERINC5 removal from the cell surface. The molecular determinants underlying this Nef function remain incompletely understood. Recent studies have found a correlation between the extent of Nef-mediated SERINC5 downregulation and the rate of disease progression. Furthermore, single-residue polymorphisms outside the known Nef functional motifs can modulate SERINC5 downregulation. The identification of a naturally occurring Nef polymorphism impairing SERINC5 downregulation in this study supports a link between Nef downregulation of SERINC5 and the rate of plasma CD4+ T cell decline. Moreover, the observed functional impairments of this polymorphism could provide clues to further elucidate unknown aspects of the SERINC5 antagonistic pathway via Nef.
Keywords: CD4; HIV; Nef; SERINC5; infectivity; membrane trafficking.
Figures






Similar articles
-
The Antagonism of HIV-1 Nef to SERINC5 Particle Infectivity Restriction Involves the Counteraction of Virion-Associated Pools of the Restriction Factor.J Virol. 2016 Nov 14;90(23):10915-10927. doi: 10.1128/JVI.01246-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27681140 Free PMC article.
-
Natural HIV-1 Nef Polymorphisms Impair SERINC5 Downregulation Activity.Cell Rep. 2019 Nov 5;29(6):1449-1457.e5. doi: 10.1016/j.celrep.2019.10.007. Cell Rep. 2019. PMID: 31693887 Free PMC article.
-
HIV-1 Nef Antagonizes SERINC5 Restriction by Downregulation of SERINC5 via the Endosome/Lysosome System.J Virol. 2018 May 14;92(11):e00196-18. doi: 10.1128/JVI.00196-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29514909 Free PMC article.
-
SERINC as a Restriction Factor to Inhibit Viral Infectivity and the Interaction with HIV.J Immunol Res. 2017;2017:1548905. doi: 10.1155/2017/1548905. Epub 2017 Nov 22. J Immunol Res. 2017. PMID: 29359168 Free PMC article. Review.
-
Role of the CD4 down-modulation activity of Nef in HIV-1 infectivity.Curr HIV Res. 2011 Oct;9(7):490-5. doi: 10.2174/157016211798842125. Curr HIV Res. 2011. PMID: 22103832 Review.
Cited by
-
Phylogenetic Reconstruction and Functional Characterization of the Ancestral Nef Protein of Primate Lentiviruses.Mol Biol Evol. 2023 Aug 3;40(8):msad164. doi: 10.1093/molbev/msad164. Mol Biol Evol. 2023. PMID: 37463439 Free PMC article.
-
The NeuroinflammatoryPotential of HIV-1 NefVariants in Modulating the Gene Expression Profile of Astrocytes.Cells. 2022 Oct 17;11(20):3256. doi: 10.3390/cells11203256. Cells. 2022. PMID: 36291123 Free PMC article.
-
Beyond Impairment of Virion Infectivity: New Activities of the Anti-HIV Host Cell Factor SERINC5.Viruses. 2024 Feb 12;16(2):284. doi: 10.3390/v16020284. Viruses. 2024. PMID: 38400059 Free PMC article. Review.
-
The role of Nef in the long-term persistence of the replication-competent HIV reservoir in South African women.J Virol. 2025 Jul 22;99(7):e0021725. doi: 10.1128/jvi.00217-25. Epub 2025 Jun 24. J Virol. 2025. PMID: 40552830 Free PMC article.
-
A Conserved Acidic Residue in the C-Terminal Flexible Loop of HIV-1 Nef Contributes to the Activity of SERINC5 and CD4 Downregulation.Viruses. 2023 Feb 28;15(3):652. doi: 10.3390/v15030652. Viruses. 2023. PMID: 36992361 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

