Biological controls for standardization and interpretation of adaptive immune receptor repertoire profiling
- PMID: 34037521
- PMCID: PMC8154019
- DOI: 10.7554/eLife.66274
Biological controls for standardization and interpretation of adaptive immune receptor repertoire profiling
Abstract
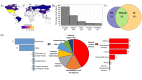
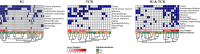
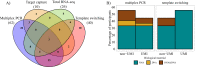
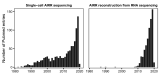
Use of adaptive immune receptor repertoire sequencing (AIRR-seq) has become widespread, providing new insights into the immune system with potential broad clinical and diagnostic applications. However, like many high-throughput technologies, it comes with several problems, and the AIRR Community was established to understand and help solve them. We, the AIRR Community's Biological Resources Working Group, have surveyed scientists about the need for standards and controls in generating and annotating AIRR-seq data. Here, we review the current status of AIRR-seq, provide the results of our survey, and based on them, offer recommendations for developing AIRR-seq standards and controls, including future work.
Keywords: B-cell Receptor (BCR); IG; T-cell Receptor (TCR); TR; antibody; immunoglobulin; immunology; inflammation; next generation sequencing (NGS).
© 2021, Trück et al.
Conflict of interest statement
JT, AE, PB, CT, DB, CS, JS, AP, ML, EM No competing interests declared, EL is consulting or is an advisor for Roche Diagnostics Corporation, Enpicom, The Antibody Society, The American Autoimmune Related Diseases Association and IEDB. AF works for Takara Bio USA, but has no ownership or stock in the company, MB During the writing of the manuscript, Magnolia Bostick was employed by Takara Bio USA, but has no ownership or stock in the company.
Figures

References
-
- Akbar R, Robert PA, Pavlović M, Jeliazkov JR, Snapkov I, Slabodkin A, Weber CR, Scheffer L, Miho E, Haff IH, Haug DTT, Lund-Johansen F, Safonova Y, Sandve GK, Greiff V. A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Reports. 2021;34:108856. doi: 10.1016/j.celrep.2021.108856. - DOI - PubMed
-
- Barennes P, Quiniou V, Shugay M, Egorov ES, Davydov AN, Chudakov DM, Uddin I, Ismail M, Oakes T, Chain B, Eugster A, Kashofer K, Rainer PP, Darko S, Ransier A, Douek DC, Klatzmann D, Mariotti-Ferrandiz E. Benchmarking of T cell receptor repertoire profiling methods reveals large systematic biases. Nature Biotechnology. 2021;39:236–245. doi: 10.1038/s41587-020-0656-3. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

