Genomic GC content drifts downward in most bacterial genomes
- PMID: 34038432
- PMCID: PMC8153448
- DOI: 10.1371/journal.pone.0244163
Genomic GC content drifts downward in most bacterial genomes
Abstract
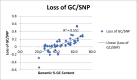
In every kingdom of life, GC->AT transitions occur more frequently than any other type of mutation due to the spontaneous deamination of cytidine. In eukaryotic genomes, this slow loss of GC base pairs is counteracted by biased gene conversion which increases genomic GC content as part of the recombination process. However, this type of biased gene conversion has not been observed in bacterial genomes, so we hypothesized that GC->AT transitions cause a reduction of genomic GC content in prokaryotic genomes on an evolutionary time scale. To test this hypothesis, we used a phylogenetic approach to analyze triplets of closely related genomes representing a wide range of the bacterial kingdom. The resulting data indicate that genomic GC content is drifting downward in bacterial genomes where GC base pairs comprise 40% or more of the total genome. In contrast, genomes containing less than 40% GC base pairs have fewer opportunities for GC->AT transitions to occur so genomic GC content is relatively stable or actually increasing. It should be noted that this observed change in genomic GC content is the net change in shared parts of the genome and does not apply to parts of the genome that have been lost or acquired since the genomes being compared shared common ancestor. However, a more detailed analysis of two Caulobacter genomes revealed that the acquisition of mobile elements by the two genomes actually reduced the total genomic GC content as well.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
GC-Content evolution in bacterial genomes: the biased gene conversion hypothesis expands.PLoS Genet. 2015 Feb 6;11(2):e1004941. doi: 10.1371/journal.pgen.1004941. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25659072 Free PMC article.
-
GC-biased gene conversion links the recombination landscape and demography to genomic base composition: GC-biased gene conversion drives genomic base composition across a wide range of species.Bioessays. 2015 Dec;37(12):1317-26. doi: 10.1002/bies.201500058. Epub 2015 Oct 7. Bioessays. 2015. PMID: 26445215 Review.
-
GC-biased gene conversion and selection affect GC content in the Oryza genus (rice).Mol Biol Evol. 2011 Sep;28(9):2695-706. doi: 10.1093/molbev/msr104. Epub 2011 Apr 18. Mol Biol Evol. 2011. PMID: 21504892
-
Impact of Recombination on the Base Composition of Bacteria and Archaea.Mol Biol Evol. 2017 Oct 1;34(10):2627-2636. doi: 10.1093/molbev/msx189. Mol Biol Evol. 2017. PMID: 28957503 Free PMC article.
-
Biased gene conversion and the evolution of mammalian genomic landscapes.Annu Rev Genomics Hum Genet. 2009;10:285-311. doi: 10.1146/annurev-genom-082908-150001. Annu Rev Genomics Hum Genet. 2009. PMID: 19630562 Review.
Cited by
-
Statistical analysis of synonymous and stop codons in pseudo-random and real sequences as a function of GC content.Sci Rep. 2023 Dec 27;13(1):22996. doi: 10.1038/s41598-023-49626-9. Sci Rep. 2023. PMID: 38151539 Free PMC article.
-
G-quadruplexes in the evolution of hepatitis B virus.Nucleic Acids Res. 2023 Aug 11;51(14):7198-7204. doi: 10.1093/nar/gkad556. Nucleic Acids Res. 2023. PMID: 37395407 Free PMC article.
-
Genomes of Novel Myxococcota Reveal Severely Curtailed Machineries for Predation and Cellular Differentiation.Appl Environ Microbiol. 2021 Nov 10;87(23):e0170621. doi: 10.1128/AEM.01706-21. Epub 2021 Sep 15. Appl Environ Microbiol. 2021. PMID: 34524899 Free PMC article.
-
Biosynthetic Gene Clusters in Sequenced Genomes of Four Contrasting Rhizobacteria in Phytopathogen Inhibition and Interaction with Capsicum annuum Roots.Microbiol Spectr. 2023 Jun 15;11(3):e0307222. doi: 10.1128/spectrum.03072-22. Epub 2023 May 24. Microbiol Spectr. 2023. PMID: 37222590 Free PMC article.
-
A simple stochastic model describing the evolution of genomic GC content in asexually reproducing organisms.Sci Rep. 2022 Nov 3;12(1):18569. doi: 10.1038/s41598-022-21709-z. Sci Rep. 2022. PMID: 36329129 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous