A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie-Wnt signaling axis in the liver
- PMID: 34038707
- PMCID: PMC8191494
- DOI: 10.1016/j.devcel.2021.05.001
A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie-Wnt signaling axis in the liver
Abstract
Single-cell transcriptomics (scRNA-seq) has revolutionized the understanding of the spatial architecture of tissue structure and function. Advancing the "transcript-centric" view of scRNA-seq analyses is presently restricted by the limited resolution of proteomics and genome-wide techniques to analyze post-translational modifications. Here, by combining spatial cell sorting with transcriptomics and quantitative proteomics/phosphoproteomics, we established the spatially resolved proteome landscape of the liver endothelium, yielding deep mechanistic insight into zonated vascular signaling mechanisms. Phosphorylation of receptor tyrosine kinases was detected preferentially in the central vein area, resulting in an atypical enrichment of tyrosine phosphorylation. Prototypic biological validation identified Tie receptor signaling as a selective and specific regulator of vascular Wnt activity orchestrating angiocrine signaling, thereby controlling hepatocyte function during liver regeneration. Taken together, the study has yielded fundamental insight into the spatial organization of liver endothelial cell signaling. Spatial sorting may be employed as a universally adaptable strategy for multiomic analyses of scRNA-seq-defined cellular (sub)-populations.
Keywords: Tie1; Tie2; Wnt; angiocrine factors; liver endothelial cell (L-EC); phosphoproteomics; proteomics; transcriptomics; vascular zonation.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


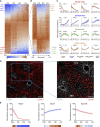
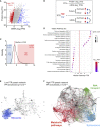

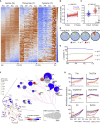

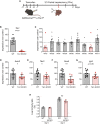
References
-
- Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: an open source software for exploring and manipulating networks. Third international AAAI conference on weblogs and social media 8, pp. 361–362.
-
- Blighe, K., Rana, S., and Lewis, M. (2020). EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. R package version 1.8.0, 10.18129/B9.bioc.EnhancedVolcano.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

