Genetic effects on liver chromatin accessibility identify disease regulatory variants
- PMID: 34038741
- PMCID: PMC8323023
- DOI: 10.1016/j.ajhg.2021.05.001
Genetic effects on liver chromatin accessibility identify disease regulatory variants
Abstract
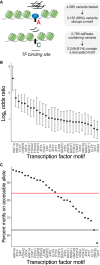
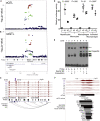
Identifying the molecular mechanisms by which genome-wide association study (GWAS) loci influence traits remains challenging. Chromatin accessibility quantitative trait loci (caQTLs) help identify GWAS loci that may alter GWAS traits by modulating chromatin structure, but caQTLs have been identified in a limited set of human tissues. Here we mapped caQTLs in human liver tissue in 20 liver samples and identified 3,123 caQTLs. The caQTL variants are enriched in liver tissue promoter and enhancer states and frequently disrupt binding motifs of transcription factors expressed in liver. We predicted target genes for 861 caQTL peaks using proximity, chromatin interactions, correlation with promoter accessibility or gene expression, and colocalization with expression QTLs. Using GWAS signals for 19 liver function and/or cardiometabolic traits, we identified 110 colocalized caQTLs and GWAS signals, 56 of which contained a predicted caPeak target gene. At the LITAF LDL-cholesterol GWAS locus, we validated that a caQTL variant showed allelic differences in protein binding and transcriptional activity. These caQTLs contribute to the epigenomic characterization of human liver and help identify molecular mechanisms and genes at GWAS loci.
Keywords: ATAC-seq; GWAS; caQTL; cardiometabolic traits; chromatin accessibility; eQTL; transcription factor motif.
Copyright © 2021 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
J.P.D. is a current employee of DNAnexus.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK093757/DK/NIDDK NIH HHS/United States
- T32 GM067553/GM/NIGMS NIH HHS/United States
- R01 DK072193/DK/NIDDK NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- T32 HL129982/HL/NHLBI NIH HHS/United States
- R25 GM055336/GM/NIGMS NIH HHS/United States
- ZIA HG000024/ImNIH/Intramural NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- R01 DK117960/DK/NIDDK NIH HHS/United States
- U01 DK105561/DK/NIDDK NIH HHS/United States
- HHSN267200700004C/DK/NIDDK NIH HHS/United States
- HHSN267200700004G/LM/NLM NIH HHS/United States
- Z01 DK070004/ImNIH/Intramural NIH HHS/United States
- T32 HL069768/HL/NHLBI NIH HHS/United States
- UM1 DK126185/DK/NIDDK NIH HHS/United States
- F31 HL146121/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

