Epigenetic regulation in Huntington's disease
- PMID: 34038804
- PMCID: PMC9110274
- DOI: 10.1016/j.neuint.2021.105074
Epigenetic regulation in Huntington's disease
Abstract
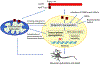
Huntington's disease (HD) is a devastating and fatal monogenic neurodegenerative disorder characterized by progressive loss of selective neurons in the brain and is caused by an abnormal expansion of CAG trinucleotide repeats in a coding exon of the huntingtin (HTT) gene. Progressive gene expression changes that begin at premanifest stages are a prominent feature of HD and are thought to contribute to disease progression. Increasing evidence suggests the critical involvement of epigenetic mechanisms in abnormal transcription in HD. Genome-wide alterations of a number of epigenetic modifications, including DNA methylation and multiple histone modifications, are associated with HD, suggesting that mutant HTT causes complex epigenetic abnormalities and chromatin structural changes, which may represent an underlying pathogenic mechanism. The causal relationship of specific epigenetic changes to early transcriptional alterations and to disease pathogenesis require further investigation. In this article, we review recent studies on epigenetic regulation in HD with a focus on DNA and histone modifications. We also discuss the contribution of epigenetic modifications to HD pathogenesis as well as potential mechanisms linking mutant HTT and epigenetic alterations. Finally, we discuss the therapeutic potential of epigenetic-based treatments.
Keywords: DNA methyltransferases (DNMTs); Epigenetic regulation; Epigenetic-based therapy; Huntington's disease (HD); Neurodegeneration; Transcription.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Figures
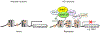
References
-
- Achour M, Le Gras S, Keime C, Parmentier F, Lejeune FX, Boutillier AL, Neri C, Davidson I, Merienne K, 2015. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum. Mol. Genet 24, 3481–3496. - PubMed
-
- Ament SA, Pearl JR, Grindeland A, St Claire J, Earls JC, Kovalenko M, Gillis T, Mysore J, Gusella JF, Lee JM, et al. , 2017. High resolution time-course mapping of early transcriptomic, molecular and cellular phenotypes in Huntington’s disease CAG knock-in mice across multiple genetic backgrounds. Hum. Mol. Genet 26, 913–922. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

