An Approach to Automatically Label and Order Brain Activity/Component Maps
- PMID: 34039009
- PMCID: PMC8867103
- DOI: 10.1089/brain.2020.0950
An Approach to Automatically Label and Order Brain Activity/Component Maps
Abstract
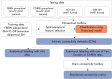
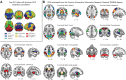
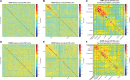
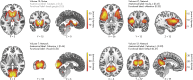
Background: Functional magnetic resonance imaging (fMRI) is a brain imaging technique that provides detailed insights into brain function and its disruption in various brain disorders. The data-driven analysis of fMRI brain activity maps involves several postprocessing steps, the first of which is identifying whether the estimated brain network maps capture signals of interest, for example, intrinsic connectivity networks (ICNs), or artifacts. This is followed by linking the ICNs to standardized anatomical and functional parcellations. Optionally, as in the study of functional network connectivity (FNC), rearranging the connectivity graph is also necessary to facilitate interpretation. Methods: Here we develop a novel and efficient method (Autolabeler) for implementing and integrating all of these processes in a fully automated manner. The Autolabeler method is pretrained on a cross-validated elastic-net regularized general linear model from the noisecloud toolbox to separate neuroscientifically meaningful ICNs from artifacts. It is capable of automatically labeling activity maps with labels from several well-known anatomical and functional parcellations. Subsequently, this method also maximizes the modularity within functional domains to generate a more systematically structured FNC matrix for post hoc network analyses. Results: Results show that our pretrained model achieves 86% accuracy at classifying ICNs from artifacts in an independent validation data set. The automatic anatomical and functional labels also have a high degree of similarity with manual labels selected by human raters. Discussion: At a time of ever-increasing rates of generating brain imaging data and analyzing brain activity, the proposed Autolabeler method is intended to automate such analyses for faster and more reproducible research. Impact statement Our proposed method is capable of implementing and integrating some of the crucial tasks in functional magnetic resonance imaging (fMRI) studies. It is the first to incorporate such tasks without the need for expert intervention. We develop an open-source toolbox for the proposed method that can function as stand-alone software and additionally provides seamless integration with the widely used group independent component analysis for fMRI toolbox (GIFT). This integration can aid investigators to conduct fMRI studies in an end-to-end automated manner.
Keywords: anatomical atlas; brain imaging; fMRI; functional network connectivity; functional parcellation.
Conflict of interest statement
No competing financial interests exist.
Figures
Similar articles
-
ICN_Atlas: Automated description and quantification of functional MRI activation patterns in the framework of intrinsic connectivity networks.Neuroimage. 2017 Dec;163:319-341. doi: 10.1016/j.neuroimage.2017.09.014. Epub 2017 Sep 9. Neuroimage. 2017. PMID: 28899742 Free PMC article.
-
Reliability and clinical utility of spatially constrained estimates of intrinsic functional networks from very short fMRI scans.Hum Brain Mapp. 2023 Apr 15;44(6):2620-2635. doi: 10.1002/hbm.26234. Epub 2023 Feb 25. Hum Brain Mapp. 2023. PMID: 36840728 Free PMC article.
-
Multi-model order spatially constrained ICA reveals highly replicable group differences and consistent predictive results from resting data: A large N fMRI schizophrenia study.Neuroimage Clin. 2023;38:103434. doi: 10.1016/j.nicl.2023.103434. Epub 2023 May 17. Neuroimage Clin. 2023. PMID: 37209635 Free PMC article.
-
Evaluation of functional MRI-based human brain parcellation: a review.J Neurophysiol. 2022 Jul 1;128(1):197-217. doi: 10.1152/jn.00411.2021. Epub 2022 Jun 8. J Neurophysiol. 2022. PMID: 35675446 Free PMC article. Review.
-
A survey on applications and analysis methods of functional magnetic resonance imaging for Alzheimer's disease.J Neurosci Methods. 2019 Apr 1;317:121-140. doi: 10.1016/j.jneumeth.2018.12.012. Epub 2018 Dec 26. J Neurosci Methods. 2019. PMID: 30593787 Review.
Cited by
-
Psychopathic traits and altered resting-state functional connectivity in incarcerated adolescent girls.Front Neuroimaging. 2023 Aug 4;2:1216494. doi: 10.3389/fnimg.2023.1216494. eCollection 2023. Front Neuroimaging. 2023. PMID: 37554634 Free PMC article.
-
Controversies and progress on standardization of large-scale brain network nomenclature.Netw Neurosci. 2023 Oct 1;7(3):864-905. doi: 10.1162/netn_a_00323. eCollection 2023. Netw Neurosci. 2023. PMID: 37781138 Free PMC article.
-
fMRI-based data-driven brain parcellation using independent component analysis.J Neurosci Methods. 2025 May;417:110403. doi: 10.1016/j.jneumeth.2025.110403. Epub 2025 Feb 18. J Neurosci Methods. 2025. PMID: 39978483
-
Data-driven approaches to neuroimaging biomarkers for neurological and psychiatric disorders: emerging approaches and examples.Curr Opin Neurol. 2021 Aug 1;34(4):469-479. doi: 10.1097/WCO.0000000000000967. Curr Opin Neurol. 2021. PMID: 34054110 Free PMC article. Review.
-
Addressing Inconsistency in Functional Neuroimaging: A Replicable Data-Driven Multi-Scale Functional Atlas for Canonical Brain Networks.bioRxiv [Preprint]. 2024 Dec 3:2024.09.09.612129. doi: 10.1101/2024.09.09.612129. bioRxiv. 2024. PMID: 39314443 Free PMC article. Preprint.
References
-
- Bell AJ, Sejnowski TJ. 1995. An information-maximization approach to blind separation and blind deconvolution. Neural Computation 7:1129–1159. - PubMed
-
- Buckner RL, Roffman JL, Smoller JW. 2014. Brain Genomics Superstruct Project (GSP). Harvard Dataverse. 10.7910/DVN/25833 Last accessed August 18, 2020. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

