Elevated type I interferon responses potentiate metabolic dysfunction, inflammation, and accelerated aging in mtDNA mutator mice
- PMID: 34039599
- PMCID: PMC8153723
- DOI: 10.1126/sciadv.abe7548
Elevated type I interferon responses potentiate metabolic dysfunction, inflammation, and accelerated aging in mtDNA mutator mice
Abstract
Mitochondrial dysfunction is a key driver of inflammatory responses in human disease. However, it remains unclear whether alterations in mitochondria-innate immune cross-talk contribute to the pathobiology of mitochondrial disorders and aging. Using the polymerase gamma (POLG) mutator model of mitochondrial DNA instability, we report that aberrant activation of the type I interferon (IFN-I) innate immune axis potentiates immunometabolic dysfunction, reduces health span, and accelerates aging in mutator mice. Mechanistically, elevated IFN-I signaling suppresses activation of nuclear factor erythroid 2-related factor 2 (NRF2), which increases oxidative stress, enhances proinflammatory cytokine responses, and accelerates metabolic dysfunction. Ablation of IFN-I signaling attenuates hyperinflammatory phenotypes by restoring NRF2 activity and reducing aerobic glycolysis, which combine to lessen cardiovascular and myeloid dysfunction in aged mutator mice. These findings further advance our knowledge of how mitochondrial dysfunction shapes innate immune responses and provide a framework for understanding mitochondria-driven immunopathology in POLG-related disorders and aging.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures
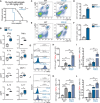
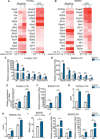
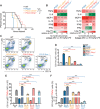
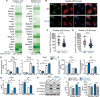
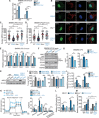

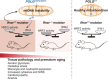
Comment in
-
Innate immunity in mutator mice.Lab Anim (NY). 2021 Jul;50(7):167. doi: 10.1038/s41684-021-00809-9. Lab Anim (NY). 2021. PMID: 34188231 No abstract available.
References
-
- West A. P., Mitochondrial dysfunction as a trigger of innate immune responses and inflammation. Toxicology 391, 54–63 (2017). - PubMed
-
- Mills E. L., Kelly B., O’Neill L. A. J., Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498 (2017). - PubMed
-
- Youle R. J., Mitochondria—Striking a balance between host and endosymbiont. Science 365, eaaw9855 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

