Treatment scheduling effects on the evolution of drug resistance in heterogeneous cancer cell populations
- PMID: 34040000
- PMCID: PMC8154902
- DOI: 10.1038/s41523-021-00270-4
Treatment scheduling effects on the evolution of drug resistance in heterogeneous cancer cell populations
Abstract
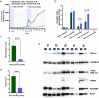
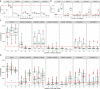
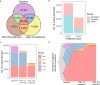
The effect of scheduling of targeted therapy combinations on drug resistance is underexplored in triple-negative breast cancer (TNBC). TNBC constitutes heterogeneous cancer cell populations the composition of which can change dynamically during treatment resulting in the selection of resistant clones with a fitness advantage. We evaluated crizotinib (ALK/MET inhibitor) and navitoclax (ABT-263; Bcl-2/Bcl-xL inhibitor) combinations in a large design consisting of 696 two-cycle sequential and concomitant treatment regimens with varying treatment dose, duration, and drug holiday length over a 26-day period in MDA-MB-231 TNBC cells and found that patterns of resistance depend on the schedule and sequence in which the drugs are given. Further, we tracked the clonal dynamics and mechanisms of resistance using DNA-integrated barcodes and single-cell RNA sequencing. Our study suggests that longer formats of treatment schedules in vitro screening assays are required to understand the effects of resistance and guide more realistically in vivo and clinical studies.
Conflict of interest statement
C.H. is currently a full-time employee of Bristol Myers Squibb. V.B.W. is currently a full-time employee of the Janssen Pharmaceutical Companies of Johnson and Johnson. L.P. has received consulting fees and honoraria from Pfizer, Astra Zeneca, Merck, Novartis, Genentech, Eisai, Pieris, Immunomedics, Seattle Genetics, Almac, Biotheranostics, and Syndax. The remaining authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

