Phylogenetic analysis of mutational robustness based on codon usage supports that the standard genetic code does not prefer extreme environments
- PMID: 34040064
- PMCID: PMC8154912
- DOI: 10.1038/s41598-021-90440-y
Phylogenetic analysis of mutational robustness based on codon usage supports that the standard genetic code does not prefer extreme environments
Abstract
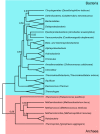
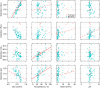
The mutational robustness of the genetic code is rarely discussed in the context of biological diversity, such as codon usage and related factors, often considered as independent of the actual organism's proteome. Here we put the living beings back to picture and use distortion as a metric of mutational robustness. Distortion estimates the expected severities of non-synonymous mutations measuring it by amino acid physicochemical properties and weighting for codon usage. Using the biological variance of codon frequencies, we interpret the mutational robustness of the standard genetic code with regards to their corresponding environments and genomic compositions (GC-content). Employing phylogenetic analyses, we show that coding fidelity in physicochemical properties can deteriorate with codon usages adapted to extreme environments and these putative effects are not the artefacts of phylogenetic bias. High temperature environments select for codon usages with decreased mutational robustness of hydrophobic, volumetric, and isoelectric properties. Selection at high saline concentrations also leads to reduced fidelity in polar and isoelectric patterns. These show that the genetic code performs best with mesophilic codon usages, strengthening the view that LUCA or its ancestors preferred lower temperature environments. Taxonomic implications, such as rooting the tree of life, are also discussed.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
The Mutational Robustness of the Genetic Code and Codon Usage in Environmental Context: A Non-Extremophilic Preference?Life (Basel). 2021 Jul 30;11(8):773. doi: 10.3390/life11080773. Life (Basel). 2021. PMID: 34440517 Free PMC article.
-
Codon Usage Optimization in the Prokaryotic Tree of Life: How Synonymous Codons Are Differentially Selected in Sequence Domains with Different Expression Levels and Degrees of Conservation.mBio. 2020 Jul 21;11(4):e00766-20. doi: 10.1128/mBio.00766-20. mBio. 2020. PMID: 32694138 Free PMC article.
-
Large-scale in silico mutagenesis experiments reveal optimization of genetic code and codon usage for protein mutational robustness.BMC Biol. 2020 Oct 20;18(1):146. doi: 10.1186/s12915-020-00870-9. BMC Biol. 2020. PMID: 33081759 Free PMC article.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
-
Codon Usage Bias: An Endless Tale.J Mol Evol. 2021 Dec;89(9-10):589-593. doi: 10.1007/s00239-021-10027-z. Epub 2021 Aug 12. J Mol Evol. 2021. PMID: 34383106 Review.
Cited by
-
The Mutational Robustness of the Genetic Code and Codon Usage in Environmental Context: A Non-Extremophilic Preference?Life (Basel). 2021 Jul 30;11(8):773. doi: 10.3390/life11080773. Life (Basel). 2021. PMID: 34440517 Free PMC article.
-
Robust genetic codes enhance protein evolvability.PLoS Biol. 2024 May 16;22(5):e3002594. doi: 10.1371/journal.pbio.3002594. eCollection 2024 May. PLoS Biol. 2024. PMID: 38754362 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

