Characterization of DNA G-Quadruplex Structures in Human Immunoglobulin Heavy Variable (IGHV) Genes
- PMID: 34040612
- PMCID: PMC8141862
- DOI: 10.3389/fimmu.2021.671944
Characterization of DNA G-Quadruplex Structures in Human Immunoglobulin Heavy Variable (IGHV) Genes
Abstract
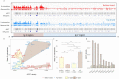
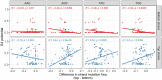
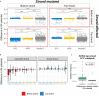
Activation-induced deaminase (AID) is a key enzyme involved in antibody diversification by initiating somatic hypermutation (SHM) and class-switch recombination (CSR) of the Immunoglobulin (Ig) loci. AID preferentially targets WRC (W=A/T, R=A/G) hotspot motifs and avoids SYC (S=C/G, Y=C/T) coldspots. G-quadruplex (G4) structures are four-stranded DNA secondary structures with key functions in transcription, translation and replication. In vitro studies have shown G4s to form and bind AID in Ig switch (S) regions. Alterations in the gene encoding AID can further disrupt AID-G4 binding and reduce CSR in vivo. However, it is still unclear whether G4s form in the variable (V) region, or how they may affect SHM. To assess the possibility of G4 formation in human V regions, we analyzed germline human Ig heavy chain V (IGHV) sequences, using a pre-trained deep learning model that predicts G4 potential. This revealed that many genes from the IGHV3 and IGHV4 families are predicted to have high G4 potential in the top and bottom strand, respectively. Different IGHV alleles also showed variability in G4 potential. Using a high-resolution (G4-seq) dataset of biochemically confirmed potential G4s in IGHV genes, we validated our computational predictions. G4-seq also revealed variation between S and V regions in the distribution of potential G4s, with the V region having overall reduced G4 abundance compared to the S region. The density of AGCT motifs, where two AGC hotspots overlap on both strands, was roughly 2.6-fold greater in the V region than the Constant (C) region, which does not mutate despite having predicted G4s at similar levels. However, AGCT motifs in both V and C regions were less abundant than in S regions. In silico mutagenesis experiments showed that G4 potentials were generally robust to mutation, although large deviations from germline states were found, mostly in framework regions. G4 potential is also associated with higher mutability of certain WRC hotspots on the same strand. In addition, CCC coldspots opposite a predicted G4 were shown to be targeted significantly more for mutation. Our overall assessment reveals plausible evidence of functional G4s forming in the Ig V region.
Keywords: G-quadruplex; IGHV genes; activation induced deaminase (AID); immunoglobulin heavy chain (Igh); somatic hypermutation (SHM).
Copyright © 2021 Tang and MacCarthy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

