Identifying Susceptibility Loci for Cutaneous Squamous Cell Carcinoma Using a Fast Sequence Kernel Association Test
- PMID: 34040636
- PMCID: PMC8141858
- DOI: 10.3389/fgene.2021.657499
Identifying Susceptibility Loci for Cutaneous Squamous Cell Carcinoma Using a Fast Sequence Kernel Association Test
Abstract
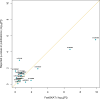
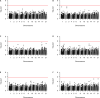
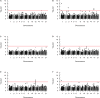
Cutaneous squamous cell carcinoma (cSCC) accounts for about 20% of all skin cancers, the most common type of malignancy in the United States. Genome-wide association studies (GWAS) have successfully identified multiple genetic variants associated with the risk of cSCC. Most of these studies were single-locus-based, testing genetic variants one-at-a-time. In this article, we performed gene-based association tests to evaluate the joint effect of multiple variants, especially rare variants, on the risk of cSCC by using a fast sequence kernel association test (fastSKAT). The study included 1,710 cSCC cases and 24,304 cancer-free controls from the Nurses' Health Study, the Nurses' Health Study II and the Health Professionals Follow-up Study. We used UCSC Genome Browser to define gene units as candidate loci, and further evaluated the association between all variants within each gene unit and disease outcome. Four genes HP1BP3, DAG1, SEPT7P2, and SLFN12 were identified using Bonferroni adjusted significance level. Our study is complementary to the existing GWASs, and our findings may provide additional insights into the etiology of cSCC. Further studies are needed to validate these findings.
Keywords: cutaneous squamous cell carcinoma; fast sequence kernel association test; generalized genetic random field; rare variants; region-based association test.
Copyright © 2021 Huang, Lyu, Li, Qureshi, Han and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-wide association studies and polygenic risk scores for skin cancer: clinically useful yet?Br J Dermatol. 2019 Dec;181(6):1146-1155. doi: 10.1111/bjd.17917. Epub 2019 Jul 7. Br J Dermatol. 2019. PMID: 30908599 Free PMC article. Review.
-
Gene expression imputation identifies candidate genes and susceptibility loci associated with cutaneous squamous cell carcinoma.Nat Commun. 2018 Oct 15;9(1):4264. doi: 10.1038/s41467-018-06149-6. Nat Commun. 2018. PMID: 30323283 Free PMC article.
-
Genetic variants in the HLA class II region associated with risk of cutaneous squamous cell carcinoma.Cancer Immunol Immunother. 2018 Jul;67(7):1123-1133. doi: 10.1007/s00262-018-2168-2. Epub 2018 May 12. Cancer Immunol Immunother. 2018. PMID: 29754218 Free PMC article.
-
Gene expression landscape of cutaneous squamous cell carcinoma progression.Br J Dermatol. 2024 Oct 17;191(5):760-774. doi: 10.1093/bjd/ljae249. Br J Dermatol. 2024. PMID: 38867481
-
Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus-based interdisciplinary guideline.Eur J Cancer. 2015 Sep;51(14):1989-2007. doi: 10.1016/j.ejca.2015.06.110. Epub 2015 Jul 25. Eur J Cancer. 2015. PMID: 26219687 Review.
Cited by
-
TGF-β/VEGF-A Genetic Variants Interplay in Genetic Susceptibility to Non-Melanocytic Skin Cancer.Genes (Basel). 2022 Jul 13;13(7):1235. doi: 10.3390/genes13071235. Genes (Basel). 2022. PMID: 35886018 Free PMC article.
References
-
- Boos D. D. (1992). On generalized score tests. Am. Stat. 46 327–333.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

