Comparative Analysis of the Circular Transcriptome in Muscle, Liver, and Testis in Three Livestock Species
- PMID: 34040640
- PMCID: PMC8141914
- DOI: 10.3389/fgene.2021.665153
Comparative Analysis of the Circular Transcriptome in Muscle, Liver, and Testis in Three Livestock Species
Abstract
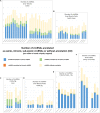
Circular RNAs have been observed in a large number of species and tissues and are now recognized as a clear component of the transcriptome. Our study takes advantage of functional datasets produced within the FAANG consortium to investigate the pervasiveness of circular RNA transcription in farm animals. We describe here the circular transcriptional landscape in pig, sheep and bovine testicular, muscular and liver tissues using total 66 RNA-seq datasets. After an exhaustive detection of circular RNAs, we propose an annotation of exonic, intronic and sub-exonic circRNAs and comparative analyses of circRNA content to evaluate the variability between individuals, tissues and species. Despite technical bias due to the various origins of the datasets, we were able to characterize some features (i) (ruminant) liver contains more exonic circRNAs than muscle (ii) in testis, the number of exonic circRNAs seems associated with the sexual maturity of the animal. (iii) a particular class of circRNAs, sub-exonic circRNAs, are produced by a large variety of multi-exonic genes (protein-coding genes, long non-coding RNAs and pseudogenes) and mono-exonic genes (protein-coding genes from mitochondrial genome and small non-coding genes). Moreover, for multi-exonic genes there seems to be a relationship between the sub-exonic circRNAs transcription level and the linear transcription level. Finally, sub-exonic circRNAs produced by mono-exonic genes (mitochondrial protein-coding genes, ribozyme, and sno) exhibit a particular behavior. Caution has to be taken regarding the interpretation of the unannotated circRNA proportion in a given tissue/species: clusters of circRNAs without annotation were characterized in genomic regions with annotation and/or assembly problems of the respective animal genomes. This study highlights the importance of improving genome annotation to better consider candidate circRNAs and to better understand the circular transcriptome. Furthermore, it emphasizes the need for considering the relative "weight" of circRNAs/parent genes for comparative analyses of several circular transcriptomes. Although there are points of agreement in the circular transcriptome of the same tissue in two species, it will be not possible to do without the characterization of it in both species.
Keywords: annotation; circular RNA; circular transcriptome; exonic circRNA; intronic circRNAs; parent genes; sub-exonic circRNA.
Copyright © 2021 Robic, Cerutti, Kühn and Faraut.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Cattle (2021). Cattle. Cambridge: Ensembl. http://www.ensembl.org/Bos_taurus/Info/Index
LinkOut - more resources
Full Text Sources
Other Literature Sources

