Timing Does Matter: Nerve-Mediated HDAC1 Paces the Temporal Expression of Morphogenic Genes During Axolotl Limb Regeneration
- PMID: 34041236
- PMCID: PMC8143519
- DOI: 10.3389/fcell.2021.641987
Timing Does Matter: Nerve-Mediated HDAC1 Paces the Temporal Expression of Morphogenic Genes During Axolotl Limb Regeneration
Abstract
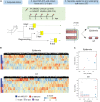
Sophisticated axolotl limb regeneration is a highly orchestrated process that requires highly regulated gene expression and epigenetic modification patterns at precise positions and timings. We previously demonstrated two waves of post-amputation expression of a nerve-mediated repressive epigenetic modulator, histone deacetylase 1 (HDAC1), at the wound healing (3 days post-amputation; 3 dpa) and blastema formation (8 dpa onward) stages in juvenile axolotls. Limb regeneration was profoundly inhibited by local injection of an HDAC inhibitor, MS-275, at the amputation sites. To explore the transcriptional response of post-amputation axolotl limb regeneration in a tissue-specific and time course-dependent manner after MS-275 treatment, we performed transcriptome sequencing of the epidermis and soft tissue (ST) at 0, 3, and 8 dpa with and without MS-275 treatment. Gene Ontology (GO) enrichment analysis of each coregulated gene cluster revealed a complex array of functional pathways in both the epidermis and ST. In particular, HDAC activities were required to inhibit the premature elevation of genes related to tissue development, differentiation, and morphogenesis. Further validation by Q-PCR in independent animals demonstrated that the expression of 5 out of 6 development- and regeneration-relevant genes that should only be elevated at the blastema stage was indeed prematurely upregulated at the wound healing stage when HDAC1 activity was inhibited. WNT pathway-associated genes were also prematurely activated under HDAC1 inhibition. Applying a WNT inhibitor to MS-275-treated amputated limbs partially rescued HDAC1 inhibition, resulting in blastema formation defects. We propose that post-amputation HDAC1 expression is at least partially responsible for pacing the expression timing of morphogenic genes to facilitate proper limb regeneration.
Keywords: axolotl limb regeneration; blastema; histone deacetylase; stage-dependent gene modulation; wound epidermis.
Copyright © 2021 Wang, Hsu, Wu, Chiou, Tsai, Lee and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Altschul SF., Gish W., Miller W., Myers EW., Lipman DJ. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

