Targeting the SARS-CoV-2-spike protein: from antibodies to miniproteins and peptides
- PMID: 34041482
- PMCID: PMC8128053
- DOI: 10.1039/d0md00385a
Targeting the SARS-CoV-2-spike protein: from antibodies to miniproteins and peptides
Abstract
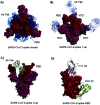
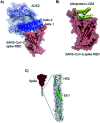
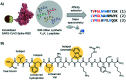
Coronavirus disease-19, caused by the novel β-coronavirus SARS-CoV-2, has created a global pandemic unseen in a century. Rapid worldwide efforts have enabled the characterization of the virus and its pathogenic mechanism. An early key finding is that SARS-CoV-2 uses spike proteins, the virus' most exposed structures, to bind to human ACE2 receptors and initiate cell invasion. Competitive targeting of the spike protein is a promising strategy to neutralize virus infectivity. This review article summarizes the discovery, binding modes and eventual applications of several classes of (bio)molecules targeting the spike protein: antibodies, nanobodies, soluble ACE2 variants, miniproteins, peptides and small molecules.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
Comparison of SARS-CoV-2 entry inhibitors based on ACE2 receptor or engineered Spike-binding peptides.J Virol. 2023 Aug 31;97(8):e0068423. doi: 10.1128/jvi.00684-23. Epub 2023 Aug 9. J Virol. 2023. PMID: 37555663 Free PMC article.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
Cited by
-
An electrochemical immunosensor using SARS-CoV-2 spike protein-nickel hydroxide nanoparticles bio-conjugate modified SPCE for ultrasensitive detection of SARS-CoV-2 antibodies.Microchem J. 2021 Nov;170:106718. doi: 10.1016/j.microc.2021.106718. Epub 2021 Aug 6. Microchem J. 2021. PMID: 34381282 Free PMC article.
-
Computational design and engineering of self-assembling multivalent microproteins with therapeutic potential against SARS-CoV-2.J Nanobiotechnology. 2024 Feb 10;22(1):58. doi: 10.1186/s12951-024-02329-3. J Nanobiotechnology. 2024. PMID: 38341574 Free PMC article.
-
In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.ACS Cent Sci. 2023 Feb 8;9(2):252-265. doi: 10.1021/acscentsci.2c01190. eCollection 2023 Feb 22. ACS Cent Sci. 2023. PMID: 36844485 Free PMC article.
-
Receptor-binding domain-anchored peptides block binding of severe acute respiratory syndrome coronavirus 2 spike proteins with cell surface angiotensin-converting enzyme 2.Front Microbiol. 2022 Sep 13;13:910343. doi: 10.3389/fmicb.2022.910343. eCollection 2022. Front Microbiol. 2022. PMID: 36177466 Free PMC article.
-
Novel Polymyxin-Inspired Peptidomimetics Targeting the SARS-CoV-2 Spike:hACE2 Interface.Int J Mol Sci. 2023 May 15;24(10):8765. doi: 10.3390/ijms24108765. Int J Mol Sci. 2023. PMID: 37240111 Free PMC article.
References
-
- Hu B. Guo H. Zhou P. Shi Z. L. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. - DOI
-
- Lu R. Zhao X. Li J. Niu P. Yang B. Wu H. Wang W. Song H. Huang B. Zhu N. Bi Y. Ma X. Zhan F. Wang L. Hu T. Zhou H. Hu Z. Zhou W. Zhao L. Chen J. Meng Y. Wang J. Lin Y. Yuan J. Xie Z. Ma J. Liu W. J. Wang D. Xu W. Holmes E. C. Gao G. F. Wu G. Chen W. Shi W. Tan W. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

