The tridecaptins: non-ribosomal peptides that selectively target Gram-negative bacteria
- PMID: 34041489
- PMCID: PMC8127968
- DOI: 10.1039/d0md00413h
The tridecaptins: non-ribosomal peptides that selectively target Gram-negative bacteria
Abstract
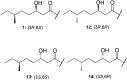
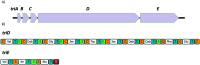
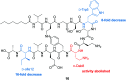
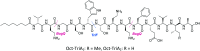
Tridecaptins are a re-emerging class of non-ribosomal antibacterial peptides (NRAPs) with potent activity against highly problematic strains of Gram-negative bacteria. An intricate mode of action has been reported to explain the bactericidal activity of these NRAPs, wherein they bind selectivity to the Gram-negative version of the peptidoglycan precursor lipid II on the outer leaflet of the inner membrane and disrupt the proton-motive force. Tridecaptins are highly amenable to synthetic modification owing to their linear structure, therefore, various synthetic analogues have been reported, several of which have enhanced antimicrobial activity, reduced cost of synthesis and/or improved stability towards d-peptidase mediated hydrolysis. It has also been demonstrated that unacylated tridecaptins can act synergistically with clinically relevant antibiotics by sensitizing the outer membrane. This review will summarize past literature on the development/discovery of novel tridecaptin analogues (up until the end of 2020), some of which may be useful therapeutic agents to treat insidious Gram-negative bacterial infections.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There is no conflict of interest to declare.
Figures







Similar articles
-
Rational design of new cyclic analogues of the antimicrobial lipopeptide tridecaptin A1.Chem Commun (Camb). 2018 Sep 25;54(75):10634-10637. doi: 10.1039/c8cc05790g. Epub 2018 Sep 4. Chem Commun (Camb). 2018. PMID: 30179243 Free PMC article.
-
Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.Proc Natl Acad Sci U S A. 2016 Oct 11;113(41):11561-11566. doi: 10.1073/pnas.1608623113. Epub 2016 Sep 29. Proc Natl Acad Sci U S A. 2016. PMID: 27688760 Free PMC article.
-
Studies of antibacterial activity (in vitro and in vivo) and mode of action for des-acyl tridecaptins (DATs).Eur J Med Chem. 2024 Feb 5;265:116097. doi: 10.1016/j.ejmech.2023.116097. Epub 2023 Dec 23. Eur J Med Chem. 2024. PMID: 38157595
-
Mechanisms of bactericidal action and resistance of polymyxins for Gram-positive bacteria.Appl Microbiol Biotechnol. 2020 May;104(9):3771-3780. doi: 10.1007/s00253-020-10525-y. Epub 2020 Mar 10. Appl Microbiol Biotechnol. 2020. PMID: 32157424 Review.
-
Lipid trafficking to the outer membrane of Gram-negative bacteria.Mol Microbiol. 2006 May;60(3):542-52. doi: 10.1111/j.1365-2958.2006.05130.x. Mol Microbiol. 2006. PMID: 16629659 Review.
Cited by
-
The Role of Antimicrobial Peptides as Antimicrobial and Antibiofilm Agents in Tackling the Silent Pandemic of Antimicrobial Resistance.Molecules. 2022 May 6;27(9):2995. doi: 10.3390/molecules27092995. Molecules. 2022. PMID: 35566343 Free PMC article. Review.
-
Systematic Determination of the Impact of Structural Edits on Peptide Accumulation into Mycobacteria.bioRxiv [Preprint]. 2025 Mar 12:2025.01.17.633618. doi: 10.1101/2025.01.17.633618. bioRxiv. 2025. Update in: ACS Chem Biol. 2025 Aug 15;20(8):1962-1979. doi: 10.1021/acschembio.5c00330. PMID: 39868157 Free PMC article. Updated. Preprint.
-
Multi-drug resistant gram-negative bacterial pneumonia: etiology, risk factors, and drug resistance patterns.Pneumonia (Nathan). 2022 May 5;14(1):4. doi: 10.1186/s41479-022-00096-z. Pneumonia (Nathan). 2022. PMID: 35509063 Free PMC article. Review.
-
A novel family of non-secreted tridecaptin lipopeptide produced by Paenibacillus elgii.Amino Acids. 2022 Nov;54(11):1477-1489. doi: 10.1007/s00726-022-03187-9. Epub 2022 Jul 21. Amino Acids. 2022. PMID: 35864259
-
Linearization of the Brevicidine and Laterocidine Lipopeptides Yields Analogues That Retain Full Antibacterial Activity.J Med Chem. 2023 Apr 27;66(8):6002-6009. doi: 10.1021/acs.jmedchem.3c00308. Epub 2023 Apr 18. J Med Chem. 2023. PMID: 37071814 Free PMC article.
References
-
- O'Neill J., Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations, 2014
-
- World Health Organization, Antibacterial Agents in Clinical Development – An analysis of the antibacterial clinical development pipeline, including tuberculosis, 2017
-
- World Health Organization, Global Tuberculosis Report, 2019
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

