Rapid and inexpensive preparation of genome-wide nucleosome footprints from model and non-model organisms
- PMID: 34041500
- PMCID: PMC8141940
- DOI: 10.1016/j.xpro.2021.100486
Rapid and inexpensive preparation of genome-wide nucleosome footprints from model and non-model organisms
Abstract
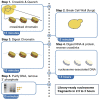
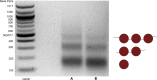
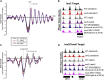
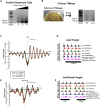
MNase-seq (micrococcal nuclease sequencing) is used to map nucleosome positions in eukaryotic genomes to study the relationship between chromatin structure and DNA-dependent processes. Current protocols require at least two days to isolate nucleosome-protected DNA fragments. We have developed a streamlined protocol for S. cerevisiae and other fungi which takes only three hours. Modified protocols were developed for wild fungi and mammalian cells. This method for rapidly producing sequencing-ready nucleosome footprints from several organisms makes MNase-seq faster and easier, with less chemical waste.
Keywords: Genomics; Model Organisms; Molecular Biology; Sequencing.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
The effect of micrococcal nuclease digestion on nucleosome positioning data.PLoS One. 2010 Dec 29;5(12):e15754. doi: 10.1371/journal.pone.0015754. PLoS One. 2010. PMID: 21206756 Free PMC article.
-
Standardized collection of MNase-seq experiments enables unbiased dataset comparisons.BMC Mol Biol. 2012 May 6;13:15. doi: 10.1186/1471-2199-13-15. BMC Mol Biol. 2012. PMID: 22559821 Free PMC article.
-
DNA Accessibility by MNase Digestions.Methods Mol Biol. 2018;1689:77-82. doi: 10.1007/978-1-4939-7380-4_7. Methods Mol Biol. 2018. PMID: 29027166
-
Genome-Wide Analysis of Nucleosome Positions, Occupancy, and Accessibility in Yeast: Nucleosome Mapping, High-Resolution Histone ChIP, and NCAM.Curr Protoc Mol Biol. 2014 Oct 1;108:21.28.1-21.28.16. doi: 10.1002/0471142727.mb2128s108. Curr Protoc Mol Biol. 2014. PMID: 25271716 Free PMC article. Review.
-
The proto-chromatosome: A fundamental subunit of chromatin?Nucleus. 2016 Jul 3;7(4):382-7. doi: 10.1080/19491034.2016.1220466. Nucleus. 2016. PMID: 27645053 Free PMC article. Review.
Cited by
-
Tup1 is critical for transcriptional repression in Quiescence in S. cerevisiae.PLoS Genet. 2022 Dec 21;18(12):e1010559. doi: 10.1371/journal.pgen.1010559. eCollection 2022 Dec. PLoS Genet. 2022. PMID: 36542663 Free PMC article.
-
The ACF chromatin-remodeling complex is essential for Polycomb repression.Elife. 2022 Mar 8;11:e77595. doi: 10.7554/eLife.77595. Elife. 2022. PMID: 35257662 Free PMC article.
-
Skeletal muscle differentiation induces wide-ranging nucleosome repositioning in muscle gene promoters.Sci Rep. 2024 Apr 24;14(1):9396. doi: 10.1038/s41598-024-60236-x. Sci Rep. 2024. PMID: 38658615 Free PMC article.
-
Osmotic disruption of chromatin induces Topoisomerase 2 activity at sites of transcriptional stress.Nat Commun. 2024 Dec 5;15(1):10606. doi: 10.1038/s41467-024-54567-6. Nat Commun. 2024. PMID: 39639049 Free PMC article.
-
Predicting the effect of CRISPR-Cas9-based epigenome editing.bioRxiv [Preprint]. 2025 Feb 28:2023.10.03.560674. doi: 10.1101/2023.10.03.560674. bioRxiv. 2025. PMID: 37873127 Free PMC article. Preprint.
References
-
- Cam H.P., Whitehall S. Micrococcal nuclease digestion of schizosaccharomyces pombe chromatin. Cold Spring Harb. Protoc. 2016;2016:996–1000. - PubMed
-
- Cole H.A., Howard B.H., Clark D.J. Genome-wide mapping of nucleosomes in yeast using paired-end sequencing. Methods Enzymol. 2012;513:145–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

