A Novel Circulating MicroRNA for the Detection of Acute Myocarditis
- PMID: 34042389
- PMCID: PMC8258773
- DOI: 10.1056/NEJMoa2003608
A Novel Circulating MicroRNA for the Detection of Acute Myocarditis
Erratum in
-
A Novel Circulating MicroRNA for the Detection of Acute Myocarditis.N Engl J Med. 2022 Nov 17;387(20):1912. doi: 10.1056/NEJMx220012. Epub 2022 Oct 24. N Engl J Med. 2022. PMID: 36378600 No abstract available.
Abstract
Background: The diagnosis of acute myocarditis typically requires either endomyocardial biopsy (which is invasive) or cardiovascular magnetic resonance imaging (which is not universally available). Additional approaches to diagnosis are desirable. We sought to identify a novel microRNA for the diagnosis of acute myocarditis.
Methods: To identify a microRNA specific for myocarditis, we performed microRNA microarray analyses and quantitative polymerase-chain-reaction (qPCR) assays in sorted CD4+ T cells and type 17 helper T (Th17) cells after inducing experimental autoimmune myocarditis or myocardial infarction in mice. We also performed qPCR in samples from coxsackievirus-induced myocarditis in mice. We then identified the human homologue for this microRNA and compared its expression in plasma obtained from patients with acute myocarditis with the expression in various controls.
Results: We confirmed that Th17 cells, which are characterized by the production of interleukin-17, are a characteristic feature of myocardial injury in the acute phase of myocarditis. The microRNA mmu-miR-721 was synthesized by Th17 cells and was present in the plasma of mice with acute autoimmune or viral myocarditis but not in those with acute myocardial infarction. The human homologue, designated hsa-miR-Chr8:96, was identified in four independent cohorts of patients with myocarditis. The area under the receiver-operating-characteristic curve for this novel microRNA for distinguishing patients with acute myocarditis from those with myocardial infarction was 0.927 (95% confidence interval, 0.879 to 0.975). The microRNA retained its diagnostic value in models after adjustment for age, sex, ejection fraction, and serum troponin level.
Conclusions: After identifying a novel microRNA in mice and humans with myocarditis, we found that the human homologue (hsa-miR-Chr8:96) could be used to distinguish patients with myocarditis from those with myocardial infarction. (Funded by the Spanish Ministry of Science and Innovation and others.).
Copyright © 2021 Massachusetts Medical Society.
Figures

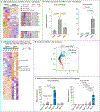

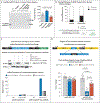

Comment in
-
Promising biomarker for the early, non-invasive diagnosis of myocarditis.Nat Rev Cardiol. 2021 Sep;18(9):612. doi: 10.1038/s41569-021-00584-7. Nat Rev Cardiol. 2021. PMID: 34103712 No abstract available.
-
Exploring RNA biomarkers in patients with acute myocarditis.Eur Heart J. 2021 Sep 14;42(35):3425-3426. doi: 10.1093/eurheartj/ehab435. Eur Heart J. 2021. PMID: 34352107 No abstract available.
-
A Novel Circulating MicroRNA for the Detection of Acute Myocarditis.N Engl J Med. 2022 Sep 29;387(13):1240. doi: 10.1056/NEJMc2115639. N Engl J Med. 2022. PMID: 36170510 No abstract available.
References
-
- Heymans S, Eriksson U, Lehtonen J, Cooper LT Jr. The quest for new approaches in myocarditis and inflammatory cardiomyopathy. J Am Coll Cardiol 2016;68: 2348–64. - PubMed
-
- Frustaci A, Russo MA, Chimenti C. Randomized study on the efficacy of immunosuppressive therapy in patients with virus-negative inflammatory cardiomyopathy: the TIMIC study. Eur Heart J 2009; 30:1995–2002. - PubMed
-
- Caforio AL, Marcolongo R, Basso C, Iliceto S. Clinical presentation and diagnosis of myocarditis. Heart 2015;101:1332–44. - PubMed
-
- Felker GM, Thompson RE, Hare JM, et al. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med 2000;342:1077–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
