Cell influx and contractile actomyosin force drive mammary bud growth and invagination
- PMID: 34042944
- PMCID: PMC8164091
- DOI: 10.1083/jcb.202008062
Cell influx and contractile actomyosin force drive mammary bud growth and invagination
Abstract
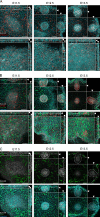
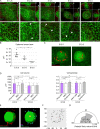
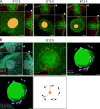
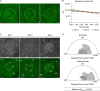
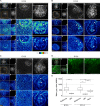
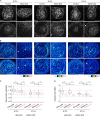
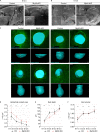
The mammary gland develops from the surface ectoderm during embryogenesis and proceeds through morphological phases defined as placode, hillock, bud, and bulb stages followed by branching morphogenesis. During this early morphogenesis, the mammary bud undergoes an invagination process where the thickened bud initially protrudes above the surface epithelium and then transforms to a bulb and sinks into the underlying mesenchyme. The signaling pathways regulating the early morphogenetic steps have been identified to some extent, but the underlying cellular mechanisms remain ill defined. Here, we use 3D and 4D confocal microscopy to show that the early growth of the mammary rudiment is accomplished by migration-driven cell influx, with minor contributions of cell hypertrophy and proliferation. We delineate a hitherto undescribed invagination mechanism driven by thin, elongated keratinocytes-ring cells-that form a contractile rim around the mammary bud and likely exert force via the actomyosin network. Furthermore, we show that conditional deletion of nonmuscle myosin IIA (NMIIA) impairs invagination, resulting in abnormal mammary bud shape.
© 2021 Trela et al.
Figures







References
-
- Agostinelli, C., and Lund U.. 2017. R package ‘circular’: Circular Statistics (version 0.4-93). https://r-forge.r-project.org/projects/circular/ (accessed March 13, 2021)
-
- Andl, T., Ahn K., Kairo A., Chu E.Y., Wine-Lee L., Reddy S.T., Croft N.J., Cebra-Thomas J.A., Metzger D., Chambon P., et al. 2004. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 131:2257–2268. 10.1242/dev.01125 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

