FGF signalling plays similar roles in development and regeneration of the skeleton in the brittle star Amphiura filiformis
- PMID: 34042967
- PMCID: PMC8180256
- DOI: 10.1242/dev.180760
FGF signalling plays similar roles in development and regeneration of the skeleton in the brittle star Amphiura filiformis
Abstract
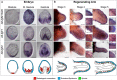
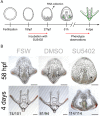
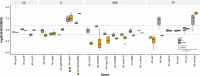
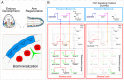
Regeneration as an adult developmental process is in many aspects similar to embryonic development. Although many studies point out similarities and differences, no large-scale, direct and functional comparative analyses between development and regeneration of a specific cell type or structure in one animal exist. Here, we use the brittle star Amphiura filiformis to characterise the role of the FGF signalling pathway during skeletal development in embryos and arm regeneration. In both processes, we find ligands expressed in ectodermal cells that flank underlying skeletal mesenchymal cells, which express the receptors. Perturbation of FGF signalling showed inhibited skeleton formation in both embryogenesis and regeneration, without affecting other key developmental processes. Differential transcriptome analysis finds mostly differentiation genes rather than transcription factors to be downregulated in both contexts. Moreover, comparative gene analysis allowed us to discover brittle star-specific differentiation genes. In conclusion, our results show that the FGF pathway is crucial for skeletogenesis in the brittle star, as in other deuterostomes, and provide evidence for the re-deployment of a developmental gene regulatory module during regeneration.
Keywords: Biomineralization; Echinoderm; Regulatory networks; Signalling; Vegf.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
-
- Altenhoff, A. M., Glover, N. M., Train, C. M., Kaleb, K., Warwick Vesztrocy, A., Dylus, D., de Farias, T. M., Zile, K., Stevenson, C., Long, J.et al. (2018). The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 46, D477-D485. 10.1093/nar/gkx1019 - DOI - PMC - PubMed
-
- Altenhoff, A. M., Levy, J., Zarowiecki, M., Vesztrocy, A. W., Dalquen, D. A., Müller, S., Swanson, B. J., Agrawal, A. D., Zucker, M., Stache-Crain, B.et al. (2019). OMA standalone: orthology inference among public and custom genomes and transcriptomes. Genome Res. 29, 1-12. 10.1101/gr.241141.118 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

