Infrared Based Saliva Screening Test for COVID-19
- PMID: 34043272
- PMCID: PMC8222893
- DOI: 10.1002/anie.202104453
Infrared Based Saliva Screening Test for COVID-19
Abstract
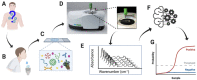
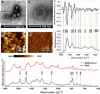
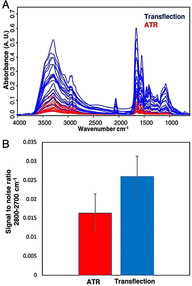
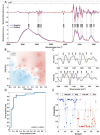
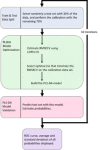
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in an unprecedented need for diagnostic testing that is critical in controlling the spread of COVID-19. We propose a portable infrared spectrometer with purpose-built transflection accessory for rapid point-of-care detection of COVID-19 markers in saliva. Initially, purified virion particles were characterized with Raman spectroscopy, synchrotron infrared (IR) and AFM-IR. A data set comprising 171 transflection infrared spectra from 29 subjects testing positive for SARS-CoV-2 by RT-qPCR and 28 testing negative, was modeled using Monte Carlo Double Cross Validation with 50 randomized test and model sets. The testing sensitivity was 93 % (27/29) with a specificity of 82 % (23/28) that included positive samples on the limit of detection for RT-qPCR. Herein, we demonstrate a proof-of-concept high throughput infrared COVID-19 test that is rapid, inexpensive, portable and utilizes sample self-collection thus minimizing the risk to healthcare workers and ideally suited to mass screening.
Keywords: COVID-19 diagnostic; Fourier transform infrared (FTIR) spectroscopy; Raman spectroscopy; SARS-CoV-2; saliva.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organisation Coronavirus Disease (COVID-19) Dashboard, W.H.O., https://covid19.who.int/ 2020.
Publication types
MeSH terms
Grants and funding
- Proposal ID. 16460 and 16476/Australian Nuclear Science and Technology Organisation
- CE140100011/Australian Research Council
- Monash Green Chemical Futures Special Project/Faculty of Science, Monash University
- 1117766/National Health and Medical Research Council of Australia
- RYC2019-026556-I/2019 Ramon y Cajal Programme
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

