Chronic T cell receptor stimulation unmasks NK receptor signaling in peripheral T cell lymphomas via epigenetic reprogramming
- PMID: 34043588
- PMCID: PMC8245185
- DOI: 10.1172/JCI139675
Chronic T cell receptor stimulation unmasks NK receptor signaling in peripheral T cell lymphomas via epigenetic reprogramming
Abstract
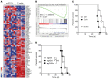
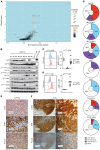
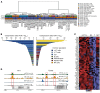
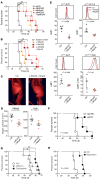
Peripheral T cell lymphomas (PTCLs) represent a significant unmet medical need with dismal clinical outcomes. The T cell receptor (TCR) is emerging as a key driver of T lymphocyte transformation. However, the role of chronic TCR activation in lymphomagenesis and in lymphoma cell survival is still poorly understood. Using a mouse model, we report that chronic TCR stimulation drove T cell lymphomagenesis, whereas TCR signaling did not contribute to PTCL survival. The combination of kinome, transcriptome, and epigenome analyses of mouse PTCLs revealed a NK cell-like reprogramming of PTCL cells with expression of NK receptors (NKRs) and downstream signaling molecules such as Tyrobp and SYK. Activating NKRs were functional in PTCLs and dependent on SYK activity. In vivo blockade of NKR signaling prolonged mouse survival, demonstrating the addiction of PTCLs to NKRs and downstream SYK/mTOR activity for their survival. We studied a large collection of human primary samples and identified several PTCLs recapitulating the phenotype described in this model by their expression of SYK and the NKR, suggesting a similar mechanism of lymphomagenesis and establishing a rationale for clinical studies targeting such molecules.
Keywords: Hematology; Immunology; Lymphomas; T cell receptor; T cells.
Conflict of interest statement
Figures




Similar articles
-
CD1d-restricted peripheral T cell lymphoma in mice and humans.J Exp Med. 2016 May 2;213(5):841-57. doi: 10.1084/jem.20150794. Epub 2016 Apr 11. J Exp Med. 2016. PMID: 27069116 Free PMC article.
-
Cytotoxic protein expression in natural killer cell lymphomas and in alpha beta and gamma delta peripheral T-cell lymphomas.J Pathol. 1997 Dec;183(4):432-9. doi: 10.1002/(SICI)1096-9896(199712)183:4<432::AID-PATH942>3.0.CO;2-4. J Pathol. 1997. PMID: 9496260
-
The fusion kinase ITK-SYK mimics a T cell receptor signal and drives oncogenesis in conditional mouse models of peripheral T cell lymphoma.J Exp Med. 2010 May 10;207(5):1031-44. doi: 10.1084/jem.20092042. Epub 2010 May 3. J Exp Med. 2010. PMID: 20439541 Free PMC article.
-
Expression of cytotoxic proteins in peripheral T-cell and natural killer-cell (NK) lymphomas: association with extranodal site, NK or Tgammadelta phenotype, anaplastic morphology and CD30 expression.Leuk Lymphoma. 2000 Jul;38(3-4):317-26. doi: 10.3109/10428190009087022. Leuk Lymphoma. 2000. PMID: 10830738 Review.
-
The Role of Adaptor Proteins in the Biology of Natural Killer T (NKT) Cells.Front Immunol. 2019 Jun 25;10:1449. doi: 10.3389/fimmu.2019.01449. eCollection 2019. Front Immunol. 2019. PMID: 31293596 Free PMC article. Review.
Cited by
-
Education and Empowering Special Forces to Eradicate Secret Defectors: Immune System-Based Treatment Approaches for Mature T- and NK-Cell Malignancies.Cancers (Basel). 2023 Apr 28;15(9):2532. doi: 10.3390/cancers15092532. Cancers (Basel). 2023. PMID: 37173999 Free PMC article. Review.
-
Mutations Affecting Genes in the Proximal T-Cell Receptor Signaling Pathway in Peripheral T-Cell Lymphoma.Cancers (Basel). 2022 Jul 29;14(15):3716. doi: 10.3390/cancers14153716. Cancers (Basel). 2022. PMID: 35954378 Free PMC article. Review.
-
Single-cell RNA sequencing comparison of CD4+, CD8+ and T-cell receptor γδ+ cutaneous T-cell lymphomas reveals subset-specific molecular phenotypes.Br J Dermatol. 2025 Jan 24;192(2):269-282. doi: 10.1093/bjd/ljae313. Br J Dermatol. 2025. PMID: 39133553 Free PMC article.
-
SOHO State of the Art Updates and Next Questions | New Pathways and New Targets in PTCL: Staying on Target.Clin Lymphoma Myeloma Leuk. 2023 Aug;23(8):561-574. doi: 10.1016/j.clml.2023.04.007. Epub 2023 Apr 19. Clin Lymphoma Myeloma Leuk. 2023. PMID: 37142534 Free PMC article. Review.
-
KIR3DL2 may represent a novel therapeutic target in aggressive systemic peripheral T-cell lymphoma.Haematologica. 2023 Oct 1;108(10):2830-2836. doi: 10.3324/haematol.2022.282220. Haematologica. 2023. PMID: 37165836 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

