Antibody Watch: Text mining antibody specificity from the literature
- PMID: 34043624
- PMCID: PMC8189493
- DOI: 10.1371/journal.pcbi.1008967
Antibody Watch: Text mining antibody specificity from the literature
Abstract
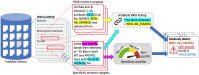
Antibodies are widely used reagents to test for expression of proteins and other antigens. However, they might not always reliably produce results when they do not specifically bind to the target proteins that their providers designed them for, leading to unreliable research results. While many proposals have been developed to deal with the problem of antibody specificity, it is still challenging to cover the millions of antibodies that are available to researchers. In this study, we investigate the feasibility of automatically generating alerts to users of problematic antibodies by extracting statements about antibody specificity reported in the literature. The extracted alerts can be used to construct an "Antibody Watch" knowledge base containing supporting statements of problematic antibodies. We developed a deep neural network system and tested its performance with a corpus of more than two thousand articles that reported uses of antibodies. We divided the problem into two tasks. Given an input article, the first task is to identify snippets about antibody specificity and classify if the snippets report that any antibody exhibits non-specificity, and thus is problematic. The second task is to link each of these snippets to one or more antibodies mentioned in the snippet. The experimental evaluation shows that our system can accurately perform the classification task with 0.925 weighted F1-score, linking with 0.962 accuracy, and 0.914 weighted F1 when combined to complete the joint task. We leveraged Research Resource Identifiers (RRID) to precisely identify antibodies linked to the extracted specificity snippets. The result shows that it is feasible to construct a reliable knowledge base about problematic antibodies by text mining.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: M.E.M, J.S.G., A.B. have an equity interest in SciCrunch, Inc., a company that may potentially benefit from the research results. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Figures
References
-
- Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

