Efficient embryonic homozygous gene conversion via RAD51-enhanced interhomolog repair
- PMID: 34043941
- PMCID: PMC8240950
- DOI: 10.1016/j.cell.2021.04.035
Efficient embryonic homozygous gene conversion via RAD51-enhanced interhomolog repair
Abstract
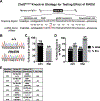
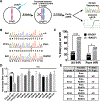
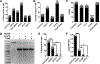
Searching for factors to improve knockin efficiency for therapeutic applications, biotechnology, and generation of non-human primate models of disease, we found that the strand exchange protein RAD51 can significantly increase Cas9-mediated homozygous knockin in mouse embryos through an interhomolog repair (IHR) mechanism. IHR is a hallmark of meiosis but only occurs at low frequencies in somatic cells, and its occurrence in zygotes is controversial. Using multiple approaches, we provide evidence for an endogenous IHR mechanism in the early embryo that can be enhanced by RAD51. This process can be harnessed to generate homozygotes from wild-type zygotes using exogenous donors and to convert heterozygous alleles into homozygous alleles without exogenous templates. Furthermore, we identify additional IHR-promoting factors and describe features of IHR events. Together, our findings show conclusive evidence for IHR in mouse embryos and describe an efficient method for enhanced gene conversion.
Keywords: BCCIP; CRISPR; DNA repair; RAD51; USP1; WDR48; gene conversion; genome editing; homologous recombination; interhomolog repair.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests MIT and J.J.W., T.A., M.W., Q.Z., and G.F. have submitted a patent application (U.S. patent application 16/260,630) related to the use of enhanced interhomolog repair for gene editing purposes.
Figures


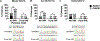

References
-
- Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, and Thomas PQ (2018). Large deletions induced by Cas9 cleavage. Nature 560, E8–E9. - PubMed
-
- Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, and Lichtenstein L (2019). Calling Somatic SNVs and Indels with Mutect2.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

