SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression
- PMID: 34043946
- PMCID: PMC8118641
- DOI: 10.1016/j.celrep.2021.109197
SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression
Abstract
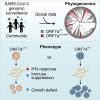
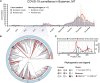
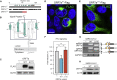
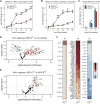
Over 950,000 whole-genome sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been determined for viruses isolated from around the world. These sequences are critical for understanding the spread and evolution of SARS-CoV-2. Using global phylogenomics, we show that mutations frequently occur in the C-terminal end of ORF7a. We isolate one of these mutant viruses from a patient sample and use viral challenge experiments to link this isolate (ORF7aΔ115) to a growth defect. ORF7a is implicated in immune modulation, and we show that the C-terminal truncation negates anti-immune activities of the protein, which results in elevated type I interferon response to the viral infection. Collectively, this work indicates that ORF7a mutations occur frequently, and that these changes affect viral mechanisms responsible for suppressing the immune response.
Keywords: IFN response; ORF7a; SARS-CoV-2.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.W. is the founder of SurGene LLC and VIRIS Detection Systems Inc. B.W., A. Nemudryi, and A. Nemudraia are inventors on patents related to CRISPR-Cas systems and applications thereof.
Figures
Update of
-
SARS-CoV-2 genomic surveillance identifies naturally occurring truncations of ORF7a that limit immune suppression.medRxiv [Preprint]. 2021 Mar 10:2021.02.22.21252253. doi: 10.1101/2021.02.22.21252253. medRxiv. 2021. Update in: Cell Rep. 2021 Jun 1;35(9):109197. doi: 10.1016/j.celrep.2021.109197. PMID: 33655280 Free PMC article. Updated. Preprint.
Similar articles
-
SARS-CoV-2 genomic surveillance identifies naturally occurring truncations of ORF7a that limit immune suppression.medRxiv [Preprint]. 2021 Mar 10:2021.02.22.21252253. doi: 10.1101/2021.02.22.21252253. medRxiv. 2021. Update in: Cell Rep. 2021 Jun 1;35(9):109197. doi: 10.1016/j.celrep.2021.109197. PMID: 33655280 Free PMC article. Updated. Preprint.
-
Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities.Cell Rep. 2021 May 18;35(7):109126. doi: 10.1016/j.celrep.2021.109126. Epub 2021 Apr 27. Cell Rep. 2021. PMID: 33974846 Free PMC article.
-
Functional landscape of SARS-CoV-2 cellular restriction.Mol Cell. 2021 Jun 17;81(12):2656-2668.e8. doi: 10.1016/j.molcel.2021.04.008. Epub 2021 Apr 13. Mol Cell. 2021. PMID: 33930332 Free PMC article.
-
SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns.Front Immunol. 2021 Jul 7;12:708264. doi: 10.3389/fimmu.2021.708264. eCollection 2021. Front Immunol. 2021. PMID: 34305949 Free PMC article. Review.
-
SARS-CoV-2 infection and the antiviral innate immune response.J Mol Cell Biol. 2020 Nov 26;12(12):963-967. doi: 10.1093/jmcb/mjaa071. J Mol Cell Biol. 2020. PMID: 33377937 Free PMC article. Review. No abstract available.
Cited by
-
PBLD enhances antiviral innate immunity by promoting the p53-USP4-MAVS signaling axis.Proc Natl Acad Sci U S A. 2024 Dec 3;121(49):e2401174121. doi: 10.1073/pnas.2401174121. Epub 2024 Nov 26. Proc Natl Acad Sci U S A. 2024. PMID: 39589880 Free PMC article.
-
SARS-CoV-2 one year on: evidence for ongoing viral adaptation.J Gen Virol. 2021 Apr;102(4):001584. doi: 10.1099/jgv.0.001584. J Gen Virol. 2021. PMID: 33855951 Free PMC article. Review.
-
The rise and spread of the SARS-CoV-2 AY.122 lineage in Russia.Virus Evol. 2022 Mar 5;8(1):veac017. doi: 10.1093/ve/veac017. eCollection 2022. Virus Evol. 2022. PMID: 35371558 Free PMC article.
-
Epidemiological associations with genomic variation in SARS-CoV-2.Sci Rep. 2021 Nov 26;11(1):23023. doi: 10.1038/s41598-021-02548-w. Sci Rep. 2021. PMID: 34837008 Free PMC article.
-
COVID-19, Obesity, and GRP78: Unraveling the Pathological Link.J Obes Metab Syndr. 2023 Sep 30;32(3):183-196. doi: 10.7570/jomes23053. J Obes Metab Syndr. 2023. PMID: 37752707 Free PMC article. Review.
References
-
- Andolfo I., Russo R., Lasorsa A.V., Cantalupo S., Rosato B.E., Bonfiglio F., Frisso G., Pasquale A., Cassese G.M., Servillo G., et al. Common variants at 21q22.3 locus influence MX1 gene expression and susceptibility to severe COVID-19. medRxiv. 2020 doi: 10.1101/2020.12.18.20248470. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

