E484K as an innovative phylogenetic event for viral evolution: Genomic analysis of the E484K spike mutation in SARS-CoV-2 lineages from Brazil
- PMID: 34044192
- PMCID: PMC8143912
- DOI: 10.1016/j.meegid.2021.104941
E484K as an innovative phylogenetic event for viral evolution: Genomic analysis of the E484K spike mutation in SARS-CoV-2 lineages from Brazil
Abstract
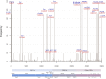
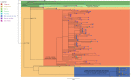
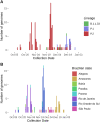
The COVID-19 pandemic caused by SARS-CoV-2 has affected millions of people since its beginning in 2019. The propagation of new lineages and the discovery of key mechanisms adopted by the virus to overlap the immune system are central topics for the entire public health policies, research and disease management. Since the second semester of 2020, the mutation E484K has been progressively found in the Brazilian territory, composing different lineages over time. It brought multiple concerns related to the risk of reinfection and the effectiveness of new preventive and treatment strategies due to the possibility of escaping from neutralizing antibodies. To better characterize the current scenario we performed genomic and phylogenetic analyses of the E484K mutated genomes sequenced from Brazilian samples in 2020. From October 2020, more than 40% of the sequenced genomes present the E484K mutation, which was identified in three different lineages (P.1, P.2 and B.1.1.33 - posteriorly renamed as N.9) in four Brazilian regions. We also evaluated the presence of E484K associated mutations and identified selective pressures acting on the spike protein, leading us to some insights about adaptive and purifying selection driving the virus evolution.
Keywords: COVID-19; E484K; Infectious diseases; Severe acute respiratory syndrome coronavirus 2; Viral evolution.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil.Emerg Infect Dis. 2021 May;27(5):1522-1524. doi: 10.3201/eid2705.210191. Epub 2021 Feb 19. Emerg Infect Dis. 2021. PMID: 33605869 Free PMC article.
-
A Potential SARS-CoV-2 Variant of Interest (VOI) Harboring Mutation E484K in the Spike Protein Was Identified within Lineage B.1.1.33 Circulating in Brazil.Viruses. 2021 Apr 21;13(5):724. doi: 10.3390/v13050724. Viruses. 2021. PMID: 33919314 Free PMC article.
-
Mutation hotspots and spatiotemporal distribution of SARS-CoV-2 lineages in Brazil, February 2020-2021.Virus Res. 2021 Oct 15;304:198532. doi: 10.1016/j.virusres.2021.198532. Epub 2021 Aug 5. Virus Res. 2021. PMID: 34363852 Free PMC article.
-
Implication of SARS-CoV-2 Immune Escape Spike Variants on Secondary and Vaccine Breakthrough Infections.Front Immunol. 2021 Nov 3;12:742167. doi: 10.3389/fimmu.2021.742167. eCollection 2021. Front Immunol. 2021. PMID: 34804022 Free PMC article. Review.
-
Recent progress on the mutations of SARS-CoV-2 spike protein and suggestions for prevention and controlling of the pandemic.Infect Genet Evol. 2021 Sep;93:104971. doi: 10.1016/j.meegid.2021.104971. Epub 2021 Jun 17. Infect Genet Evol. 2021. PMID: 34146731 Free PMC article. Review.
Cited by
-
Genomic and virologic characterization of samples from a shipboard outbreak of COVID-19 reveals distinct variants within limited temporospatial parameters.Front Microbiol. 2022 Aug 10;13:960932. doi: 10.3389/fmicb.2022.960932. eCollection 2022. Front Microbiol. 2022. PMID: 36033872 Free PMC article.
-
Evolutionary history of the SARS-CoV-2 Gamma variant of concern (P.1): a perfect storm.Genet Mol Biol. 2022 Mar 9;45(1):e20210309. doi: 10.1590/1678-4685-GMB-2021-0309. eCollection 2022. Genet Mol Biol. 2022. PMID: 35266951 Free PMC article.
-
Evolutionary Traits and Genomic Surveillance of SARS-CoV-2 in South America.Glob Health Epidemiol Genom. 2022 May 18;2022:8551576. doi: 10.1155/2022/8551576. eCollection 2022. Glob Health Epidemiol Genom. 2022. PMID: 35655960 Free PMC article. Review.
-
Estimation of Secondary Household Attack Rates for Emergent Spike L452R Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants Detected by Genomic Surveillance at a Community-Based Testing Site in San Francisco.Clin Infect Dis. 2022 Jan 7;74(1):32-39. doi: 10.1093/cid/ciab283. Clin Infect Dis. 2022. PMID: 33788923 Free PMC article.
-
Insights into the mutation T1117I in the spike and the lineage B.1.1.389 of SARS-CoV-2 circulating in Costa Rica.Gene Rep. 2022 Jun;27:101554. doi: 10.1016/j.genrep.2022.101554. Epub 2022 Feb 8. Gene Rep. 2022. PMID: 35155843 Free PMC article.
References
-
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., Manna R., Casa E., Hyseni I., Benincasa L., Montomoli E., Amaro R.E., McLellan J.S., Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 2020.12.28.424451. - PMC - PubMed
-
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. - PMC - PubMed
-
- Chi X., Yan R., Zhang Jun, Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang Jinlong, Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

