A glycolysis-based three-gene signature predicts survival in patients with lung squamous cell carcinoma
- PMID: 34044809
- PMCID: PMC8161559
- DOI: 10.1186/s12885-021-08360-z
A glycolysis-based three-gene signature predicts survival in patients with lung squamous cell carcinoma
Abstract
Background: Lung cancer is one of the most lethal and most prevalent malignant tumors worldwide, and lung squamous cell carcinoma (LUSC) is one of the major histological subtypes. Although numerous biomarkers have been found to be associated with prognosis in LUSC, the prediction effect of a single gene biomarker is insufficient, especially for glycolysis-related genes. Therefore, we aimed to develop a novel glycolysis-related gene signature to predict survival in patients with LUSC.
Methods: The mRNA expression files and LUSC clinical information were obtained from The Cancer Genome Atlas (TCGA) dataset.
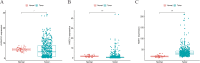
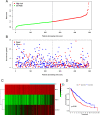
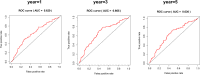
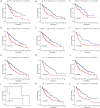
Results: Based on Gene Set Enrichment Analysis (GSEA), we found 5 glycolysis-related gene sets that were significantly enriched in LUSC tissues. Univariate and multivariate Cox proportional regression models were performed to choose prognostic-related gene signatures. Based on a Cox proportional regression model, a risk score for a three-gene signature (HKDC1, ALDH7A1, and MDH1) was established to divide patients into high-risk and low-risk subgroups. Multivariate Cox regression analysis indicated that the risk score for this three-gene signature can be used as an independent prognostic indicator in LUSC. Additionally, based on the cBioPortal database, the rate of genomic alterations in the HKDC1, ALDH7A1, and MDH1 genes were 1.9, 1.1, and 5% in LUSC patients, respectively.
Conclusion: A glycolysis-based three-gene signature could serve as a novel biomarker in predicting the prognosis of patients with LUSC and it also provides additional gene targets that can be used to cure LUSC patients.
Keywords: Gene signature; Glycolysis; Lung cancer; Prognosis.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


References
-
- Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52(Pt 1):103–109. doi: 10.1016/j.semcancer.2017.11.019. - DOI - PMC - PubMed
-
- Molinier O, Goupil F, Debieuvre D, Auliac JB, Jeandeau S, Lacroix S, Martin F, Grivaux M. Five-year survival and prognostic factors according to histology in 6101 non-small-cell lung cancer patients. Respir Med Res. 2019;77:46–54. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

