Chromosomal copy number heterogeneity predicts survival rates across cancers
- PMID: 34045449
- PMCID: PMC8160133
- DOI: 10.1038/s41467-021-23384-6
Chromosomal copy number heterogeneity predicts survival rates across cancers
Abstract
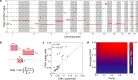
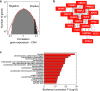
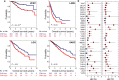
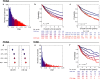
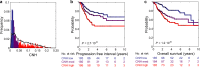
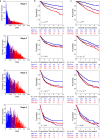
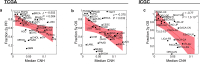
Survival rates of cancer patients vary widely within and between malignancies. While genetic aberrations are at the root of all cancers, individual genomic features cannot explain these distinct disease outcomes. In contrast, intra-tumour heterogeneity (ITH) has the potential to elucidate pan-cancer survival rates and the biology that drives cancer prognosis. Unfortunately, a comprehensive and effective framework to measure ITH across cancers is missing. Here, we introduce a scalable measure of chromosomal copy number heterogeneity (CNH) that predicts patient survival across cancers. We show that the level of ITH can be derived from a single-sample copy number profile. Using gene-expression data and live cell imaging we demonstrate that ongoing chromosomal instability underlies the observed heterogeneity. Analysing 11,534 primary cancer samples from 37 different malignancies, we find that copy number heterogeneity can be accurately deduced and predicts cancer survival across tissues of origin and stages of disease. Our results provide a unifying molecular explanation for the different survival rates observed between cancer types.
Conflict of interest statement
E.v.D., B.Y., L.V. and D.M.M. are listed as inventors in a pending patent application (NL82151) filed by Oncode Institute on behalf of the Academisch Medisch Centrum, covering the content of the paper. The remaining authors declare no competing interests.
Figures

Similar articles
-
Novel insights into breast cancer copy number genetic heterogeneity revealed by single-cell genome sequencing.Elife. 2020 May 13;9:e51480. doi: 10.7554/eLife.51480. Elife. 2020. PMID: 32401198 Free PMC article.
-
An integrative pan-cancer investigation reveals common genetic and transcriptional alterations of AMPK pathway genes as important predictors of clinical outcomes across major cancer types.BMC Cancer. 2020 Aug 17;20(1):773. doi: 10.1186/s12885-020-07286-2. BMC Cancer. 2020. PMID: 32807122 Free PMC article.
-
Pan-cancer analysis of the extent and consequences of intratumor heterogeneity.Nat Med. 2016 Jan;22(1):105-13. doi: 10.1038/nm.3984. Epub 2015 Nov 30. Nat Med. 2016. PMID: 26618723 Free PMC article.
-
Resolving genetic heterogeneity in cancer.Nat Rev Genet. 2019 Jul;20(7):404-416. doi: 10.1038/s41576-019-0114-6. Nat Rev Genet. 2019. PMID: 30918367 Review.
-
Intra-tumour heterogeneity - going beyond genetics.FEBS J. 2016 Jun;283(12):2245-58. doi: 10.1111/febs.13705. Epub 2016 Apr 1. FEBS J. 2016. PMID: 26945550 Review.
Cited by
-
Combined Focused Next-Generation Sequencing Assays to Guide Precision Oncology in Solid Tumors: A Retrospective Analysis from an Institutional Molecular Tumor Board.Cancers (Basel). 2022 Sep 12;14(18):4430. doi: 10.3390/cancers14184430. Cancers (Basel). 2022. PMID: 36139590 Free PMC article.
-
Chromosomal instability in aneuploid acute lymphoblastic leukemia associates with disease progression.EMBO Mol Med. 2024 Jan;16(1):64-92. doi: 10.1038/s44321-023-00006-w. Epub 2023 Dec 15. EMBO Mol Med. 2024. PMID: 38177531 Free PMC article.
-
Prognostic Value of Molecular Intratumor Heterogeneity in Primary Oral Cancer and Its Lymph Node Metastases Assessed by Mass Spectrometry Imaging.Molecules. 2022 Aug 25;27(17):5458. doi: 10.3390/molecules27175458. Molecules. 2022. PMID: 36080226 Free PMC article.
-
Molecular landscape and functional characterization of centrosome amplification in ovarian cancer.Nat Commun. 2023 Oct 16;14(1):6505. doi: 10.1038/s41467-023-41840-3. Nat Commun. 2023. PMID: 37845213 Free PMC article.
-
Molecular characterization of colorectal cancer related peritoneal metastatic disease.Nat Commun. 2022 Aug 4;13(1):4443. doi: 10.1038/s41467-022-32198-z. Nat Commun. 2022. PMID: 35927254 Free PMC article.
References
-
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature578, 82–93, 10.1038/s41586-020-1969-6 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

