Evolution of opposing regulatory interactions underlies the emergence of eukaryotic cell cycle checkpoints
- PMID: 34045495
- PMCID: PMC8159995
- DOI: 10.1038/s41598-021-90384-3
Evolution of opposing regulatory interactions underlies the emergence of eukaryotic cell cycle checkpoints
Abstract
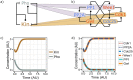
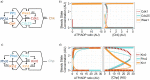
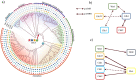
In eukaryotes the entry into mitosis is initiated by activation of cyclin-dependent kinases (CDKs), which in turn activate a large number of protein kinases to induce all mitotic processes. The general view is that kinases are active in mitosis and phosphatases turn them off in interphase. Kinases activate each other by cross- and self-phosphorylation, while phosphatases remove these phosphate groups to inactivate kinases. Crucial exceptions to this general rule are the interphase kinase Wee1 and the mitotic phosphatase Cdc25. Together they directly control CDK in an opposite way of the general rule of mitotic phosphorylation and interphase dephosphorylation. Here we investigate why this opposite system emerged and got fixed in almost all eukaryotes. Our results show that this reversed action of a kinase-phosphatase pair, Wee1 and Cdc25, on CDK is particularly suited to establish a stable G2 phase and to add checkpoints to the cell cycle. We show that all these regulators appeared together in LECA (Last Eukaryote Common Ancestor) and co-evolved in eukaryotes, suggesting that this twist in kinase-phosphatase regulation was a crucial step happening at the emergence of eukaryotes.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Phosphorylation of CDC25C by AMP-activated protein kinase mediates a metabolic checkpoint during cell-cycle G2/M-phase transition.J Biol Chem. 2018 Apr 6;293(14):5185-5199. doi: 10.1074/jbc.RA117.001379. Epub 2018 Feb 21. J Biol Chem. 2018. PMID: 29467227 Free PMC article.
-
Regulatory dephosphorylation of CDK at G₂/M in plants: yeast mitotic phosphatase cdc25 induces cytokinin-like effects in transgenic tobacco morphogenesis.Ann Bot. 2011 May;107(7):1071-86. doi: 10.1093/aob/mcr016. Epub 2011 Feb 20. Ann Bot. 2011. PMID: 21339187 Free PMC article.
-
A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit.Cell. 2011 Nov 11;147(4):803-14. doi: 10.1016/j.cell.2011.09.047. Cell. 2011. PMID: 22078879
-
Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells.Plant Mol Biol. 2000 Aug;43(5-6):595-605. doi: 10.1023/a:1006412413671. Plant Mol Biol. 2000. PMID: 11089863 Review.
-
Checking out the G(2)/M transition.Biochim Biophys Acta. 2001 May 28;1519(1-2):1-12. doi: 10.1016/s0167-4781(01)00204-4. Biochim Biophys Acta. 2001. PMID: 11406266 Review.
Cited by
-
Multiwell-based G0-PCC assay for radiation biodosimetry.Sci Rep. 2024 Aug 26;14(1):19789. doi: 10.1038/s41598-024-69243-4. Sci Rep. 2024. PMID: 39187542 Free PMC article.
-
A comprehensive review of computational cell cycle models in guiding cancer treatment strategies.NPJ Syst Biol Appl. 2024 Jul 5;10(1):71. doi: 10.1038/s41540-024-00397-7. NPJ Syst Biol Appl. 2024. PMID: 38969664 Free PMC article. Review.
-
Molecular Mechanism of Overcoming Host Resistance by the Target of Rapamycin Gene in Leptographium qinlingensis.Microorganisms. 2022 Feb 24;10(3):503. doi: 10.3390/microorganisms10030503. Microorganisms. 2022. PMID: 35336079 Free PMC article.
References
-
- Alberts B, et al. Part I introduction to the cell: cells and genomes. Mol. Biol. Cell. 2017 doi: 10.1201/9781315735368-1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

