A hybrid modeling approach for assessing mechanistic models of small molecule partitioning in vivo using a machine learning-integrated modeling platform
- PMID: 34045592
- PMCID: PMC8160209
- DOI: 10.1038/s41598-021-90637-1
A hybrid modeling approach for assessing mechanistic models of small molecule partitioning in vivo using a machine learning-integrated modeling platform
Abstract
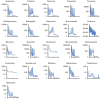
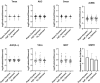
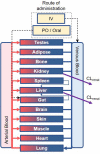
Prediction of the first-in-human dosing regimens is a critical step in drug development and requires accurate quantitation of drug distribution. Traditional in vivo studies used to characterize clinical candidate's volume of distribution are error-prone, time- and cost-intensive and lack reproducibility in clinical settings. The paper demonstrates how a computational platform integrating machine learning optimization with mechanistic modeling can be used to simulate compound plasma concentration profile and predict tissue-plasma partition coefficients with high accuracy by varying the lipophilicity descriptor logP. The approach applied to chemically diverse small molecules resulted in comparable geometric mean fold-errors of 1.50 and 1.63 in pharmacokinetic outputs for direct tissue:plasma partition and hybrid logP optimization, with the latter enabling prediction of tissue permeation that can be used to guide toxicity and efficacy dosing in human subjects. The optimization simulations required to achieve these results were parallelized on the AWS cloud and generated outputs in under 5 h. Accuracy, speed, and scalability of the framework indicate that it can be used to assess the relevance of other mechanistic relationships implicated in pharmacokinetic-pharmacodynamic phenomena with a lower risk of overfitting datasets and generate large database of physiologically-relevant drug disposition for further integration with machine learning models.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

