Retinoic acid signaling is critical during the totipotency window in early mammalian development
- PMID: 34045724
- PMCID: PMC8195742
- DOI: 10.1038/s41594-021-00590-w
Retinoic acid signaling is critical during the totipotency window in early mammalian development
Erratum in
-
Author Correction: Retinoic acid signaling is critical during the totipotency window in early mammalian development.Nat Struct Mol Biol. 2022 Mar;29(3):282. doi: 10.1038/s41594-022-00740-8. Nat Struct Mol Biol. 2022. PMID: 35165459 Free PMC article. No abstract available.
Abstract
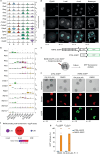
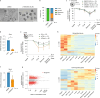
Totipotent cells hold enormous potential for regenerative medicine. Thus, the development of cellular models recapitulating totipotent-like features is of paramount importance. Cells resembling the totipotent cells of early embryos arise spontaneously in mouse embryonic stem (ES) cell cultures. Such '2-cell-like-cells' (2CLCs) recapitulate 2-cell-stage features and display expanded cell potential. Here, we used 2CLCs to perform a small-molecule screen to identify new pathways regulating the 2-cell-stage program. We identified retinoids as robust inducers of 2CLCs and the retinoic acid (RA)-signaling pathway as a key component of the regulatory circuitry of totipotent cells in embryos. Using single-cell RNA-seq, we reveal the transcriptional dynamics of 2CLC reprogramming and show that ES cells undergo distinct cellular trajectories in response to RA. Importantly, endogenous RA activity in early embryos is essential for zygotic genome activation and developmental progression. Overall, our data shed light on the gene regulatory networks controlling cellular plasticity and the totipotency program.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Ishiuchi, T. & Torres-Padilla, M.-E. Towards an understanding of the regulatory mechanisms of totipotency. Curr. Opin. Genet. Dev.23, 512–518 (2013). - PubMed
-
- Wu, G. & Schöler, H. R. Lineage segregation in the totipotent embryo. Curr. Top. Dev. Biol.117, 301–317 (2016). - PubMed
-
- Tarkowski, A. K. Experiments on the development of isolated blastomeres of mouse eggs. Nature184, 1286–1287 (1959). - PubMed
-
- Togashi, M. Production of monozygotic twins by splitting of 2-cell stage embryos in mice. Jpn J. Anim. Reprod.33, 51–57 (1987).
-
- Sotomaru, Y., Kato, Y. & Tsunoda, Y. Production of monozygotic twins after freezing and thawing of bisected mouse embryos. Cryobiology37, 139–145 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

