Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer
- PMID: 34045764
- PMCID: PMC8483261
- DOI: 10.1038/s41588-021-00874-3
Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer
Abstract
Ionizing radiation causes DNA damage and is a mainstay for cancer treatment, but understanding of its genomic impact is limited. We analyzed mutational spectra following radiotherapy in 190 paired primary and recurrent gliomas from the Glioma Longitudinal Analysis Consortium and 3,693 post-treatment metastatic tumors from the Hartwig Medical Foundation. We identified radiotherapy-associated significant increases in the burden of small deletions (5-15 bp) and large deletions (20+ bp to chromosome-arm length). Small deletions were characterized by a larger span size, lacking breakpoint microhomology and were genomically more dispersed when compared to pre-existing deletions and deletions in non-irradiated tumors. Mutational signature analysis implicated classical non-homologous end-joining-mediated DNA damage repair and APOBEC mutagenesis following radiotherapy. A high radiation-associated deletion burden was associated with worse clinical outcomes, suggesting that effective repair of radiation-induced DNA damage is detrimental to patient survival. These results may be leveraged to predict sensitivity to radiation therapy in recurrent cancer.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
R.G.W.V. is a co-founder of Boundless Bio, Inc., which was not involved in the research presented here. R.G.W.V. is a member of Scientific Advisory Board of the Hartwig Medical Foundation. F.P.B. has performed consulting for Bristol Myers Squibb.
R.G.W.V., E.K., K.J.A., and F.P.B. are listed as inventors on a patent application filed by The Jackson Laboratory, related to the findings described here. The remaining authors declare no competing interests.
Figures






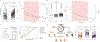
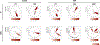

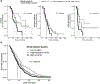
References
-
- Barton MB et al.Estimating the demand for radiotherapy from the evidence: a review of changes from 2003 to 2012. Radiother Oncol 112, 140–4 (2014). - PubMed
-
- Tyldesley S et al.Estimating the need for radiotherapy for patients with prostate, breast, and lung cancers: verification of model estimates of need with radiotherapy utilization data from British Columbia. Int J Radiat Oncol Biol Phys 79, 1507–15 (2011). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

