A second generation of 1,2,4-oxadiazole derivatives with enhanced solubility for inhibition of 3-hydroxykynurenine transaminase (HKT) from Aedes aegypti
- PMID: 34046611
- PMCID: PMC8127416
- DOI: 10.1039/d0md00305k
A second generation of 1,2,4-oxadiazole derivatives with enhanced solubility for inhibition of 3-hydroxykynurenine transaminase (HKT) from Aedes aegypti
Abstract
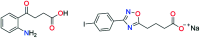
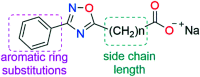
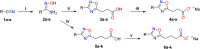
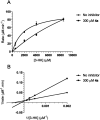
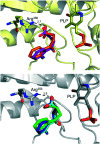
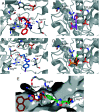
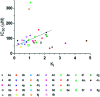
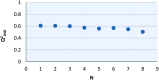
The most widely used method for the control of the Aedes aegypti mosquito population is the chemical control method. It represents a time- and cost-effective way to curb several diseases (e.g. dengue, Zika, chikungunya, yellow fever) through vector control. For this reason, the discovery of new compounds with a distinct mode of action from the available ones is essential in order to minimize the rise of insecticide resistance. Detoxification enzymes are an attractive target for the discovery of new insecticides. The kynurenine pathway is an important metabolic pathway, and it leads to the chemically stable xanthurenic acid, biosynthesized from 3-hydroxykynurenine, a precursor of reactive oxygen and nitrogen species, by the enzyme 3-hydroxykynurenine transaminase (HKT). Previously, we have reported the effectiveness of 1,2,4-oxadiazole derivatives acting as larvicides for A. aegypti and AeHKT inhibitors from in vitro and in silico studies. Here, we report the synthesis of new sodium 4-[3-(aryl)-1,2,4-oxadiazol-5-yl] propanoates and the cognate HKT-inhibitory activity. These new derivatives act as competitive inhibitors with IC50 values in the range of 42 to 339 μM. We further performed molecular docking simulations and QSAR analysis for the previously synthesized sodium 4-[3-(aryl)-1,2,4-oxadiazol-5-yl] butanoates reported earlier by our group and the data produced herein. Most of the 1,2,4-oxadiazole derivatives, including the canonical compounds for both series, showed a similar binding mode with HKT. The binding occurs similarly to the co-crystallized inhibitor via anchoring to Arg356 and positioning of the aromatic ring and its substituents outwards at the entry of the active site. QSAR analysis was performed in search of more than 770 molecular descriptors to establish a relationship between the lowest energy conformations and the IC50 values. The five best descriptors were selected to create and validate the model, which exhibited parameters that attested to its robustness and predictability. In summary, we observed that compounds with a para substitution and heavier groups (i.e. CF3 and NO2 substituents) had an enhanced HKT-inhibition profile. These compounds comprise a series described as AeHKT inhibitors via enzymatic inhibition experiments, opening the way to further the development of new substances with higher potency against HKT from Aedes aegypti.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
Similar articles
-
Inhibition of 3-Hydroxykynurenine Transaminase from Aedes aegypti and Anopheles gambiae: A Mosquito-Specific Target to Combat the Transmission of Arboviruses.ACS Bio Med Chem Au. 2023 Feb 16;3(2):211-222. doi: 10.1021/acsbiomedchemau.2c00080. eCollection 2023 Apr 19. ACS Bio Med Chem Au. 2023. PMID: 37101811 Free PMC article.
-
Discovery of 1,2,4-oxadiazole derivatives as a novel class of noncompetitive inhibitors of 3-hydroxykynurenine transaminase (HKT) from Aedes aegypti.Bioorg Med Chem. 2020 Jan 15;28(2):115252. doi: 10.1016/j.bmc.2019.115252. Epub 2019 Dec 9. Bioorg Med Chem. 2020. PMID: 31864777
-
Fast and low-cost evaluation of hydroxykynurenine activity.MethodsX. 2020 Jul 2;7:100982. doi: 10.1016/j.mex.2020.100982. eCollection 2020. MethodsX. 2020. PMID: 32685382 Free PMC article.
-
In silico models for predicting vector control chemicals targeting Aedes aegypti.SAR QSAR Environ Res. 2014;25(10):805-35. doi: 10.1080/1062936X.2014.958291. Epub 2014 Oct 2. SAR QSAR Environ Res. 2014. PMID: 25275884 Free PMC article. Review.
-
Pyrethroid resistance in the dengue vector Aedes aegypti in Southeast Asia: present situation and prospects for management.Parasit Vectors. 2018 Jun 4;11(1):332. doi: 10.1186/s13071-018-2899-0. Parasit Vectors. 2018. PMID: 29866193 Free PMC article. Review.
Cited by
-
Inhibition of 3-Hydroxykynurenine Transaminase from Aedes aegypti and Anopheles gambiae: A Mosquito-Specific Target to Combat the Transmission of Arboviruses.ACS Bio Med Chem Au. 2023 Feb 16;3(2):211-222. doi: 10.1021/acsbiomedchemau.2c00080. eCollection 2023 Apr 19. ACS Bio Med Chem Au. 2023. PMID: 37101811 Free PMC article.
-
Biochemical Evolution of a Potent Target of Mosquito Larvicide, 3-Hydroxykynurenine Transaminase.Molecules. 2022 Aug 2;27(15):4929. doi: 10.3390/molecules27154929. Molecules. 2022. PMID: 35956879 Free PMC article.
-
Novel Hydroxamic Acids Containing Aryl-Substituted 1,2,4- or 1,3,4-Oxadiazole Backbones and an Investigation of Their Antibiotic Potentiation Activity.Int J Mol Sci. 2023 Dec 20;25(1):96. doi: 10.3390/ijms25010096. Int J Mol Sci. 2023. PMID: 38203266 Free PMC article.
-
Molecular docking and molecular dynamics simulation studies of inhibitor candidates against Anopheles gambiae 3-hydroxykynurenine transaminase and implications on vector control.Heliyon. 2025 Jan 2;11(1):e41633. doi: 10.1016/j.heliyon.2025.e41633. eCollection 2025 Jan 15. Heliyon. 2025. PMID: 39866405 Free PMC article.
References
-
- Rossini G., Landini M. P. and Sambri V., in Chikungunya Virus: Methods and Protocols, ed. J. J. H. Chu and S. K. Ang, Springer Science+Business Media, New york, 2016, vol. 1426, pp. 3–10 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

