Targeting fibroblast growth factor receptors to combat aggressive ependymoma
- PMID: 34046693
- PMCID: PMC8270873
- DOI: 10.1007/s00401-021-02327-x
Targeting fibroblast growth factor receptors to combat aggressive ependymoma
Abstract
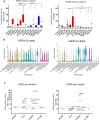
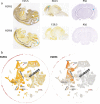
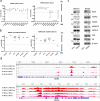
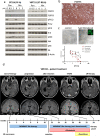
Ependymomas (EPN) are central nervous system tumors comprising both aggressive and more benign molecular subtypes. However, therapy of the high-risk subtypes posterior fossa group A (PF-A) and supratentorial RELA-fusion positive (ST-RELA) is limited to gross total resection and radiotherapy, as effective systemic treatment concepts are still lacking. We have recently described fibroblast growth factor receptors 1 and 3 (FGFR1/FGFR3) as oncogenic drivers of EPN. However, the underlying molecular mechanisms and their potential as therapeutic targets have not yet been investigated in detail. Making use of transcriptomic data across 467 EPN tissues, we found that FGFR1 and FGFR3 were both widely expressed across all molecular groups. FGFR3 mRNA levels were enriched in ST-RELA showing the highest expression among EPN as well as other brain tumors. We further identified high expression levels of fibroblast growth factor 1 and 2 (FGF1, FGF2) across all EPN subtypes while FGF9 was elevated in ST-EPN. Interrogation of our EPN single-cell RNA-sequencing data revealed that FGFR3 was further enriched in cycling and progenitor-like cell populations. Corroboratively, we found FGFR3 to be predominantly expressed in radial glia cells in both mouse embryonal and human brain datasets. Moreover, we detected alternative splicing of the FGFR1/3-IIIc variant, which is known to enhance ligand affinity and FGFR signaling. Dominant-negative interruption of FGFR1/3 activation in PF-A and ST-RELA cell models demonstrated inhibition of key oncogenic pathways leading to reduced cell growth and stem cell characteristics. To explore the feasibility of therapeutically targeting FGFR, we tested a panel of FGFR inhibitors in 12 patient-derived EPN cell models revealing sensitivity in the low-micromolar to nano-molar range. Finally, we gain the first clinical evidence for the activity of the FGFR inhibitor nintedanib in the treatment of a patient with recurrent ST-RELA. Together, these preclinical and clinical data suggest FGFR inhibition as a novel and feasible approach to combat aggressive EPN.
Keywords: Brain tumor; Ependymoma; FGFR; Pediatric cancer; Small molecule inhibitors.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Arabzade A, Zhao Y, Varadharajan S, Chen H-C, Jessa S, Rivas B, et al. ZFTA-RELA dictates oncogenic transcriptional programs to drive aggressive supratentorial ependymoma. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1066. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

